An optimized TALEN application for mutagenesis and screening in Drosophila melanogaster
- PMID: 26196022
- PMCID: PMC4501208
- DOI: 10.1080/21592799.2015.1023423
An optimized TALEN application for mutagenesis and screening in Drosophila melanogaster
Abstract
Transcription activator-like effector nucleases (TALENs) emerged as powerful tools for locus-specific genome engineering. Due to the ease of TALEN assembly, the key to streamlining TALEN-induced mutagenesis lies in identifying efficient TALEN pairs and optimizing TALEN mRNA injection concentrations to minimize the effort to screen for mutant offspring. Here we present a simple methodology to quantitatively assess bi-allelic TALEN cutting, as well as approaches that permit accurate measures of somatic and germline mutation rates in Drosophila melanogaster. We report that percent lethality from pilot injection of candidate TALEN mRNAs into Lig4 null embryos can be used to effectively gauge bi-allelic TALEN cutting efficiency and occurs in a dose-dependent manner. This timely Lig4-dependent embryonic survival assay also applies to CRISPR/Cas9-mediated _targeting. Moreover, the somatic mutation rate of individual G0 flies can be rapidly quantitated using SURVEYOR nuclease and capillary electrophoresis, and germline transmission rate determined by scoring progeny of G0 outcrosses. Together, these optimized methods provide an effective step-wise guide for routine TALEN-mediated gene editing in the fly.
Keywords: TALEN; TALENs, Transcription activator-like effector nucleases; TALEs, TAL effectors; ZFNs, Zinc Finger Nucleases; CRISPR, Clustered Regularly Interspersed Short Palindromic Repeats; Cas9, CRISPR-associated; RVDs, repeat-variable diresidues; DSBs, double-stranded breaks; NHEJ, non-homologous end joining; HR, homologous recombination; RFLP, restriction fragment length polymorphism; HRMA, high resolution melt analysis.; engineered endonuclease; genome engineering; mutagenesis; screening.
Figures
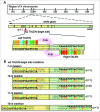
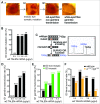
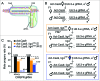
Similar articles
-
TALEN-Mediated Homologous Recombination Produces Site-Directed DNA Base Change and Herbicide-Resistant Rice.J Genet Genomics. 2016 May 20;43(5):297-305. doi: 10.1016/j.jgg.2016.03.005. Epub 2016 Mar 22. J Genet Genomics. 2016. PMID: 27180265
-
Characteristic and inheritance analysis of _targeted mutagenesis mediated by genome editing in rice.Yi Chuan. 2016 Aug;38(8):746-55. doi: 10.16288/j.yczz.16-052. Yi Chuan. 2016. PMID: 27531613
-
Silencing of end-joining repair for efficient site-specific gene insertion after TALEN/CRISPR mutagenesis in Aedes aegypti.Proc Natl Acad Sci U S A. 2015 Mar 31;112(13):4038-43. doi: 10.1073/pnas.1502370112. Epub 2015 Mar 16. Proc Natl Acad Sci U S A. 2015. PMID: 25775608 Free PMC article.
-
Genome editing with engineered nucleases in plants.Plant Cell Physiol. 2015 Mar;56(3):389-400. doi: 10.1093/pcp/pcu170. Epub 2014 Nov 20. Plant Cell Physiol. 2015. PMID: 25416289 Review.
-
Applications of Alternative Nucleases in the Age of CRISPR/Cas9.Int J Mol Sci. 2017 Nov 29;18(12):2565. doi: 10.3390/ijms18122565. Int J Mol Sci. 2017. PMID: 29186020 Free PMC article. Review.
Cited by
-
Robust CRISPR/Cas9-Mediated Tissue-Specific Mutagenesis Reveals Gene Redundancy and Perdurance in Drosophila.Genetics. 2019 Feb;211(2):459-472. doi: 10.1534/genetics.118.301736. Epub 2018 Nov 30. Genetics. 2019. PMID: 30504366 Free PMC article.
-
CRISPR/Cas9-mediated efficient white genome editing in the black soldier fly Hermetia illucens.Mol Genet Genomics. 2024 Feb 5;299(1):5. doi: 10.1007/s00438-023-02088-0. Mol Genet Genomics. 2024. PMID: 38315256
-
Mini-Review Regarding the Applicability of Genome Editing Techniques Developed for Studying Infertility.Diagnostics (Basel). 2021 Feb 5;11(2):246. doi: 10.3390/diagnostics11020246. Diagnostics (Basel). 2021. PMID: 33562517 Free PMC article. Review.
-
A Simple Method To Detect Point Mutations in Aspergillus fumigatus cyp51A Gene Using a Surveyor Nuclease Assay.Antimicrob Agents Chemother. 2020 Mar 24;64(4):e02271-19. doi: 10.1128/AAC.02271-19. Print 2020 Mar 24. Antimicrob Agents Chemother. 2020. PMID: 32015034 Free PMC article.
References
-
- Beumer K, Bhattacharyya G, Bibikova M, Trautman JK, Carroll D. Efficient gene _targeting in Drosophila with zinc-finger nucleases. Genetics 2006; 172:2391-403; PMID:16452139; http://dx.doi.org/10.1534/genetics.105.052829 - DOI - PMC - PubMed
-
- Beumer KJ, Trautman JK, Mukherjee K, Carroll D. Donor DNA Utilization during Gene _targeting with Zinc-finger Nucleases. G3 (Bethesda) 2013; 3:657-64; PMID:23550125; http://dx.doi.org/10.1534/g3.112.005439 - DOI - PMC - PubMed
-
- Liu J, Li C, Yu Z, Huang P, Wu H, Wei C, Zhu N, Shen Y, Chen Y, Zhang B, et al. . Efficient and specific modifications of the Drosophila genome by means of an easy TALEN strategy. J Genet Genomics 2012; 39:209-15; PMID:22624882; http://dx.doi.org/10.1016/j.jgg.2012.04.003 - DOI - PubMed
-
- Kondo T, Sakuma T, Wada H, Akimoto-Kato A, Yamamoto T, Hayashi S. TALEN-induced gene knock out in Drosophila. Dev Growth Differ 2014; 56:86-91; PMID:24172335; http://dx.doi.org/10.1111/dgd.12097 - DOI - PubMed
-
- Beumer KJ, Trautman JK, Christian M, Dahlem TJ, Lake CM, Hawley RS, Grunwald DJ, Voytas DF, Carroll D. Comparing zinc finger nucleases and transcription activator-like effector nucleases for gene _targeting in Drosophila. G3 (Bethesda) 2013; 3:1717-25; PMID:23979928; http://dx.doi.org/full_text - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
