Importance of reference gene selection for articular cartilage mechanobiology studies
- PMID: 26585242
- PMCID: PMC4819451
- DOI: 10.1016/j.joca.2015.11.007
Importance of reference gene selection for articular cartilage mechanobiology studies
Abstract
Objective: Identification of genes differentially expressed in mechano-biological pathways in articular cartilage provides insight into the molecular mechanisms behind initiation and/or progression of osteoarthritis (OA). Quantitative PCR (qPCR) is commonly used to measure gene expression, and is reliant on the use of reference genes for normalisation. Appropriate validation of reference gene stability is imperative for accurate data analysis and interpretation. This study determined in vitro reference gene stability in articular cartilage explants and primary chondrocytes subjected to different compressive loads and tensile strain, respectively.
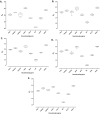
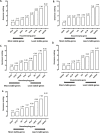
Design: The expression of eight commonly used reference genes (18s, ACTB, GAPDH, HPRT1, PPIA, RPL4, SDHA and YWHAZ) was determined by qPCR and data compared using four software packages (comparative delta-Ct method, geNorm, NormFinder and BestKeeper). Calculation of geometric means of the ranked weightings was carried out using RefFinder.
Results: Appropriate reference gene(s) for normalisation of mechanically-regulated transcript levels in articular cartilage tissue or isolated chondrocytes were dependent on experimental set-up. SDHA, YWHAZ and RPL4 were the most stable genes whilst glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and to a lesser extent Hypoxanthine-guanine phosphoribosyltransferase (HPRT), showed variable expression in response to load, demonstrating their unsuitability in such in vitro studies. The effect of using unstable reference genes to normalise the expression of aggrecan (ACAN) and matrix metalloproteinase 3 (MMP3) resulted in inaccurate quantification of these mechano-sensitive genes and erroneous interpretation/conclusions.
Conclusion: This study demonstrates that commonly used 'reference genes' may be unsuitable for in vitro cartilage chondrocyte mechanobiology studies, reinforcing the principle that careful validation of reference genes is essential prior to each experiment to obtain robust and reproducible qPCR data for analysis/interpretation.
Keywords: Articular cartilage; Chondrocytes; Mechanobiology; Quantitative PCR; Reference genes.
Copyright © 2015 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures




Similar articles
-
Selection of optimal reference genes for quantitative RT-PCR studies of boar spermatozoa cryopreservation.Cryobiology. 2014 Feb;68(1):113-21. doi: 10.1016/j.cryobiol.2014.01.004. Epub 2014 Jan 16. Cryobiology. 2014. PMID: 24440873
-
Selection of reference genes for gene expression studies in pig tissues using SYBR green qPCR.BMC Mol Biol. 2007 Aug 15;8:67. doi: 10.1186/1471-2199-8-67. BMC Mol Biol. 2007. PMID: 17697375 Free PMC article.
-
Selection of reference genes for quantitative real-time polymerase chain reaction in porcine embryos.Reprod Fertil Dev. 2017 Feb;29(2):357-367. doi: 10.1071/RD14393. Reprod Fertil Dev. 2017. PMID: 26293544
-
Effects of shear stress on articular chondrocyte metabolism.Biorheology. 2000;37(1-2):95-107. Biorheology. 2000. PMID: 10912182 Review.
-
Tissue-engineered versus native cartilage: linkage between cellular mechano-transduction and biomechanical properties.Novartis Found Symp. 2003;249:52-64; discussion 64-9, 170-4, 239-41. Novartis Found Symp. 2003. PMID: 12708649 Review.
Cited by
-
Local Tensile Stress in the Development of Posttraumatic Osteoarthritis.Biomed Res Int. 2018 Nov 4;2018:4210353. doi: 10.1155/2018/4210353. eCollection 2018. Biomed Res Int. 2018. PMID: 30519575 Free PMC article.
-
Inflammatory and degenerative phases resulting from anterior cruciate rupture in a non-invasive murine model of post-traumatic osteoarthritis.J Orthop Res. 2018 Feb 17;36(8):2118-27. doi: 10.1002/jor.23872. Online ahead of print. J Orthop Res. 2018. PMID: 29453795 Free PMC article.
-
Evaluation of reference genes for gene expression studies in mouse and N2a cell ischemic stroke models using quantitative real-time PCR.BMC Neurosci. 2018 Feb 1;19(1):3. doi: 10.1186/s12868-018-0403-6. BMC Neurosci. 2018. PMID: 29390963 Free PMC article.
-
Regulation of microRNA-221, -222, -21 and -27 in articular cartilage subjected to abnormal compressive forces.J Physiol. 2021 Jan;599(1):143-155. doi: 10.1113/JP279810. Epub 2020 Oct 31. J Physiol. 2021. PMID: 33052608 Free PMC article.
-
Development of male-larger sexual size dimorphism in a lizard: IGF1 peak long after sexual maturity overlaps with pronounced growth in males.Front Physiol. 2022 Aug 10;13:917460. doi: 10.3389/fphys.2022.917460. eCollection 2022. Front Physiol. 2022. PMID: 36035474 Free PMC article.
References
-
- Mow V.C., Ratcliffe A., Poole A.R. Cartilage and diarthrodial joints as paradigms for hierarchical materials and structures. Biomaterials. 1992;13:67–97. - PubMed
-
- Bustin S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. - PubMed
-
- Lee C.R., Grad S., Maclean J.J., Iatridis J.C., Alini M. Effect of mechanical loading on mRNA levels of common endogenous controls in articular chondrocytes and intervertebral disk. Anal Biochem. 2005;341:372–375. - PubMed
-
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

