NBR1 enables autophagy-dependent focal adhesion turnover
- PMID: 26903539
- PMCID: PMC4772495
- DOI: 10.1083/jcb.201503075
NBR1 enables autophagy-dependent focal adhesion turnover
Abstract
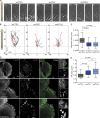
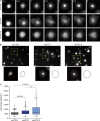
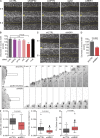
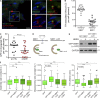
Autophagy is a catabolic pathway involving the sequestration of cellular contents into a double-membrane vesicle, the autophagosome. Although recent studies have demonstrated that autophagy supports cell migration, the underlying mechanisms remain unknown. Using live-cell imaging, we uncover that autophagy promotes optimal migratory rate and facilitates the dynamic assembly and disassembly of cell-matrix focal adhesions (FAs), which is essential for efficient motility. Additionally, our studies reveal that autophagosomes associate with FAs primarily during disassembly, suggesting autophagy locally facilitates the destabilization of cell-matrix contact sites. Furthermore, we identify the selective autophagy cargo receptor neighbor of BRCA1 (NBR1) as a key mediator of autophagy-dependent FA remodeling. NBR1 depletion impairs FA turnover and decreases _targeting of autophagosomes to FAs, whereas ectopic expression of autophagy-competent, but not autophagy-defective, NBR1 enhances FA disassembly and reduces FA lifetime during migration. Our findings provide mechanistic insight into how autophagy promotes migration by revealing a requirement for NBR1-mediated selective autophagy in enabling FA disassembly in motile cells.
© 2016 Kenific et al.
Figures


Similar articles
-
Autophagy promotes cell motility by driving focal adhesion turnover.Autophagy. 2016 Oct 2;12(10):1685-1686. doi: 10.1080/15548627.2016.1212791. Epub 2016 Aug 2. Autophagy. 2016. PMID: 27483986 Free PMC article.
-
NBR1-dependent selective autophagy is required for efficient cell-matrix adhesion site disassembly.Autophagy. 2016 Oct 2;12(10):1958-1959. doi: 10.1080/15548627.2016.1212789. Epub 2016 Aug 2. Autophagy. 2016. PMID: 27484104 Free PMC article.
-
Autophagy in adhesion and migration.J Cell Sci. 2016 Oct 15;129(20):3685-3693. doi: 10.1242/jcs.188490. Epub 2016 Sep 26. J Cell Sci. 2016. PMID: 27672021 Free PMC article. Review.
-
Revealing the three dimensional architecture of focal adhesion components to explain Ca2+-mediated turnover of focal adhesions.Biochim Biophys Acta Gen Subj. 2017 Mar;1861(3):624-635. doi: 10.1016/j.bbagen.2017.01.002. Epub 2017 Jan 5. Biochim Biophys Acta Gen Subj. 2017. PMID: 28063985
-
Mechanisms of FA-Phagy, a New Form of Selective Autophagy/Organellophagy.Front Cell Dev Biol. 2021 Dec 7;9:799123. doi: 10.3389/fcell.2021.799123. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34950664 Free PMC article. Review.
Cited by
-
Autophagy Reduces the Degradation and Promotes Membrane Localization of Occludin to Enhance the Intestinal Epithelial Tight Junction Barrier against Paracellular Macromolecule Flux.J Crohns Colitis. 2023 Apr 3;17(3):433-449. doi: 10.1093/ecco-jcc/jjac148. J Crohns Colitis. 2023. PMID: 36219473 Free PMC article.
-
THSD1 Suppresses Autophagy-Mediated Focal Adhesion Turnover by Modulating the FAK-Beclin 1 Pathway.Int J Mol Sci. 2024 Feb 10;25(4):2139. doi: 10.3390/ijms25042139. Int J Mol Sci. 2024. PMID: 38396816 Free PMC article.
-
Autophagy promotes cell motility by driving focal adhesion turnover.Autophagy. 2016 Oct 2;12(10):1685-1686. doi: 10.1080/15548627.2016.1212791. Epub 2016 Aug 2. Autophagy. 2016. PMID: 27483986 Free PMC article.
-
Autophagy Promotes Focal Adhesion Disassembly and Cell Motility of Metastatic Tumor Cells through the Direct Interaction of Paxillin with LC3.Cell Rep. 2016 May 24;15(8):1660-72. doi: 10.1016/j.celrep.2016.04.065. Epub 2016 May 12. Cell Rep. 2016. PMID: 27184837 Free PMC article.
-
Autophagy Induced by Toll-like Receptor Ligands Regulates Antigen Extraction and Presentation by B Cells.Cells. 2022 Dec 1;11(23):3883. doi: 10.3390/cells11233883. Cells. 2022. PMID: 36497137 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

