USP7 is a SUMO deubiquitinase essential for DNA replication
- PMID: 26950370
- PMCID: PMC4869841
- DOI: 10.1038/nsmb.3185
USP7 is a SUMO deubiquitinase essential for DNA replication
Abstract
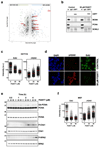
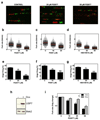
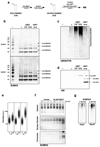
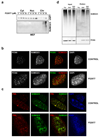
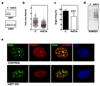
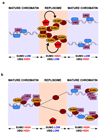
Post-translational modification of proteins by ubiquitin (Ub) and Ub-like modifiers regulates DNA replication. We have previously shown that chromatin around replisomes is rich in SUMO and poor in Ub, whereas mature chromatin exhibits an opposite pattern. How this SUMO-rich, Ub-poor environment is maintained at sites of DNA replication in mammalian cells remains unexplored. Here we identify USP7 as a replisome-enriched SUMO deubiquitinase that is essential for DNA replication. By acting on SUMO and SUMOylated proteins, USP7 counteracts their ubiquitination. Inhibition or genetic deletion of USP7 leads to the accumulation of Ub on SUMOylated proteins, which are displaced away from replisomes. Our findings provide a model explaining the differential accumulation of SUMO and Ub at replication forks and identify an essential role of USP7 in DNA replication that should be considered in the development of USP7 inhibitors as anticancer agents.
Figures

Similar articles
-
A SUMO and ubiquitin code coordinates protein traffic at replication factories.Bioessays. 2016 Dec;38(12):1209-1217. doi: 10.1002/bies.201600129. Epub 2016 Sep 26. Bioessays. 2016. PMID: 27667742 Review.
-
USP7 and VCPFAF1 define the SUMO/Ubiquitin landscape at the DNA replication fork.Cell Rep. 2021 Oct 12;37(2):109819. doi: 10.1016/j.celrep.2021.109819. Cell Rep. 2021. PMID: 34644576 Free PMC article.
-
Revealing USP7 Deubiquitinase Substrate Specificity by Unbiased Synthesis of Ubiquitin Tagged SUMO2.Biochemistry. 2020 Oct 13;59(40):3796-3801. doi: 10.1021/acs.biochem.0c00701. Epub 2020 Oct 2. Biochemistry. 2020. PMID: 33006472
-
USP7/HAUSP: A SUMO deubiquitinase at the heart of DNA replication.Bioessays. 2016 Sep;38(9):863-8. doi: 10.1002/bies.201600096. Epub 2016 Jul 4. Bioessays. 2016. PMID: 27374980 Review.
-
Coordinating DNA Replication and Mitosis through Ubiquitin/SUMO and CDK1.Int J Mol Sci. 2021 Aug 16;22(16):8796. doi: 10.3390/ijms22168796. Int J Mol Sci. 2021. PMID: 34445496 Free PMC article. Review.
Cited by
-
Advances in Deubiquitinating Enzyme Inhibition and Applications in Cancer Therapeutics.Cancers (Basel). 2020 Jun 15;12(6):1579. doi: 10.3390/cancers12061579. Cancers (Basel). 2020. PMID: 32549302 Free PMC article. Review.
-
A feedforward circuit shaped by ECT2 and USP7 contributes to breast carcinogenesis.Theranostics. 2020 Aug 29;10(23):10769-10790. doi: 10.7150/thno.46878. eCollection 2020. Theranostics. 2020. PMID: 32929379 Free PMC article.
-
The Role of Deubiquitinating Enzyme in Head and Neck Squamous Cell Carcinoma.Int J Mol Sci. 2022 Dec 29;24(1):552. doi: 10.3390/ijms24010552. Int J Mol Sci. 2022. PMID: 36613989 Free PMC article. Review.
-
The Ubiquitin-Specific Protease Usp7, a Novel Merkel Cell Polyomavirus Large T-Antigen Interaction Partner, Modulates Viral DNA Replication.J Virol. 2020 Feb 14;94(5):e01638-19. doi: 10.1128/JVI.01638-19. Print 2020 Feb 14. J Virol. 2020. PMID: 31801860 Free PMC article.
-
Mechanisms of DNA Damage Tolerance: Post-Translational Regulation of PCNA.Genes (Basel). 2018 Dec 24;10(1):10. doi: 10.3390/genes10010010. Genes (Basel). 2018. PMID: 30586904 Free PMC article. Review.
References
-
- Lehmann AR. Ubiquitin-family modifications in the replication of DNA damage. FEBS Lett. 2011;585:2772–9. - PubMed
-
- Zhang W, Qin Z, Zhang X, Xiao W. Roles of sequential ubiquitination of PCNA in DNA-damage tolerance. FEBS Lett. 2011;585:2786–94. - PubMed
-
- Huang TT, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–47. - PubMed
-
- Cohn MA, et al. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell. 2007;28:786–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

