Drugging the undruggables: exploring the ubiquitin system for drug development
- PMID: 27002218
- PMCID: PMC4822129
- DOI: 10.1038/cr.2016.31
Drugging the undruggables: exploring the ubiquitin system for drug development
Abstract
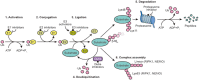
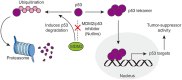
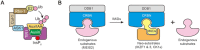
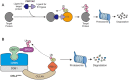
Dynamic modulation of protein levels is tightly controlled in response to physiological cues. In mammalian cells, much of the protein degradation is carried out by the ubiquitin-proteasome system (UPS). Similar to kinases, components of the ubiquitin system are often dysregulated, leading to a variety of diseases, including cancer and neurodegeneration, making them attractive drug _targets. However, so far there are only a handful of drugs _targeting the ubiquitin system that have been approved by the FDA. Here, we review possible therapeutic intervention nodes in the ubiquitin system, analyze the challenges, and highlight the most promising strategies to _target the UPS.
Figures

Similar articles
-
_targeting the ubiquitin proteasome system in haematological malignancies.Blood Rev. 2013 Nov;27(6):297-304. doi: 10.1016/j.blre.2013.10.002. Epub 2013 Oct 19. Blood Rev. 2013. PMID: 24183816 Review.
-
[Ubiquitin-proteasome pathway as a _target for therapeutic strategies].Postepy Biochem. 2017;63(4):287-303. Postepy Biochem. 2017. PMID: 29374430 Review. Polish.
-
Hidden _targets of ubiquitin proteasome system: To prevent diabetic nephropathy.Pharmacol Res. 2017 Jun;120:170-179. doi: 10.1016/j.phrs.2017.03.024. Epub 2017 Mar 29. Pharmacol Res. 2017. PMID: 28363724 Review.
-
Alternative UPS drug _targets upstream the 26S proteasome.Int J Biochem Cell Biol. 2008;40(6-7):1126-40. doi: 10.1016/j.biocel.2007.11.021. Epub 2007 Dec 8. Int J Biochem Cell Biol. 2008. PMID: 18203645 Review.
-
_targeting the ubiquitin proteasome system: beyond proteasome inhibition.Curr Pharm Des. 2013;19(22):4053-93. doi: 10.2174/1381612811319220014. Curr Pharm Des. 2013. PMID: 23181575 Review.
Cited by
-
Enzymatic Logic of Ubiquitin Chain Assembly.Front Physiol. 2019 Jul 5;10:835. doi: 10.3389/fphys.2019.00835. eCollection 2019. Front Physiol. 2019. PMID: 31333493 Free PMC article. Review.
-
A patent review of the ubiquitin ligase system: 2015-2018.Expert Opin Ther Pat. 2018 Dec;28(12):919-937. doi: 10.1080/13543776.2018.1549229. Epub 2018 Nov 23. Expert Opin Ther Pat. 2018. PMID: 30449221 Free PMC article. Review.
-
Ubiquitination in Scleroderma Fibrosis and Its Treatment.Front Immunol. 2018 Oct 17;9:2383. doi: 10.3389/fimmu.2018.02383. eCollection 2018. Front Immunol. 2018. PMID: 30386338 Free PMC article. Review.
-
Writing Histone Monoubiquitination in Human Malignancy-The Role of RING Finger E3 Ubiquitin Ligases.Genes (Basel). 2019 Jan 18;10(1):67. doi: 10.3390/genes10010067. Genes (Basel). 2019. PMID: 30669413 Free PMC article. Review.
-
Detailed Dissection of UBE3A-Mediated DDI1 Ubiquitination.Front Physiol. 2019 May 3;10:534. doi: 10.3389/fphys.2019.00534. eCollection 2019. Front Physiol. 2019. PMID: 31130875 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

