Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes
- PMID: 27029739
- PMCID: PMC4821875
- DOI: 10.1038/ncomms11089
Blood-based biomarkers of age-associated epigenetic changes in human islets associate with insulin secretion and diabetes
Abstract
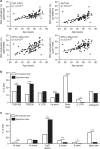
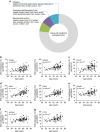
Aging associates with impaired pancreatic islet function and increased type 2 diabetes (T2D) risk. Here we examine whether age-related epigenetic changes affect human islet function and if blood-based epigenetic biomarkers reflect these changes and associate with future T2D. We analyse DNA methylation genome-wide in islets from 87 non-diabetic donors, aged 26-74 years. Aging associates with increased DNA methylation of 241 sites. These sites cover loci previously associated with T2D, for example, KLF14. Blood-based epigenetic biomarkers reflect age-related methylation changes in 83 genes identified in human islets (for example, KLF14, FHL2, ZNF518B and FAM123C) and some associate with insulin secretion and T2D. DNA methylation correlates with islet expression of multiple genes, including FHL2, ZNF518B, GNPNAT1 and HLTF. Silencing these genes in β-cells alter insulin secretion. Together, we demonstrate that blood-based epigenetic biomarkers reflect age-related DNA methylation changes in human islets, and associate with insulin secretion in vivo and T2D.
Figures



Comment in
-
Epigenetics: Blood-based markers for T2DM.Nat Rev Endocrinol. 2016 Jun;12(6):311. doi: 10.1038/nrendo.2016.63. Epub 2016 Apr 22. Nat Rev Endocrinol. 2016. PMID: 27109784 No abstract available.
Similar articles
-
Whole-Genome Bisulfite Sequencing of Human Pancreatic Islets Reveals Novel Differentially Methylated Regions in Type 2 Diabetes Pathogenesis.Diabetes. 2017 Apr;66(4):1074-1085. doi: 10.2337/db16-0996. Epub 2017 Jan 4. Diabetes. 2017. PMID: 28052964
-
Genome-wide DNA methylation analysis of human pancreatic islets from type 2 diabetic and non-diabetic donors identifies candidate genes that influence insulin secretion.PLoS Genet. 2014 Mar 6;10(3):e1004160. doi: 10.1371/journal.pgen.1004160. eCollection 2014 Mar. PLoS Genet. 2014. PMID: 24603685 Free PMC article.
-
Effects of palmitate on genome-wide mRNA expression and DNA methylation patterns in human pancreatic islets.BMC Med. 2014 Jun 23;12:103. doi: 10.1186/1741-7015-12-103. BMC Med. 2014. PMID: 24953961 Free PMC article.
-
Epigenetic regulation of insulin action and secretion - role in the pathogenesis of type 2 diabetes.J Intern Med. 2020 Aug;288(2):158-167. doi: 10.1111/joim.13049. Epub 2020 May 3. J Intern Med. 2020. PMID: 32363639 Review.
-
Does epigenetic dysregulation of pancreatic islets contribute to impaired insulin secretion and type 2 diabetes?Biochem Cell Biol. 2015 Oct;93(5):511-21. doi: 10.1139/bcb-2015-0057. Epub 2015 Aug 4. Biochem Cell Biol. 2015. PMID: 26369706 Review.
Cited by
-
Derivation and Validation of a Prediction Model for Predicting the 5-Year Incidence of Type 2 Diabetes in Non-Obese Adults: A Population-Based Cohort Study.Diabetes Metab Syndr Obes. 2021 May 11;14:2087-2101. doi: 10.2147/DMSO.S304994. eCollection 2021. Diabetes Metab Syndr Obes. 2021. PMID: 34007195 Free PMC article.
-
Making sense of the ageing methylome.Nat Rev Genet. 2022 Oct;23(10):585-605. doi: 10.1038/s41576-022-00477-6. Epub 2022 May 2. Nat Rev Genet. 2022. PMID: 35501397 Review.
-
DNA Methylation in Gestational Diabetes and its Predictive Value for Postpartum Glucose Disturbances.J Clin Endocrinol Metab. 2022 Sep 28;107(10):2748-2757. doi: 10.1210/clinem/dgac462. J Clin Endocrinol Metab. 2022. PMID: 35914803 Free PMC article.
-
Myc Is Required for Adaptive β-Cell Replication in Young Mice but Is Not Sufficient in One-Year-Old Mice Fed With a High-Fat Diet.Diabetes. 2019 Oct;68(10):1934-1949. doi: 10.2337/db18-1368. Epub 2019 Jul 10. Diabetes. 2019. PMID: 31292135 Free PMC article.
-
Epigenetic control of β-cell function and failure.Diabetes Res Clin Pract. 2017 Jan;123:24-36. doi: 10.1016/j.diabres.2016.11.009. Epub 2016 Nov 21. Diabetes Res Clin Pract. 2017. PMID: 27918975 Free PMC article. Review.
References
-
- Chang A. M. & Halter J. B. Aging and insulin secretion. Am. J. Physiol. Endocrinol. Metab. 284, E7–12 (2003). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

