Comparative Proteomic Analysis Reveals Differential Root Proteins in Medicago sativa and Medicago truncatula in Response to Salt Stress
- PMID: 27066057
- PMCID: PMC4814493
- DOI: 10.3389/fpls.2016.00424
Comparative Proteomic Analysis Reveals Differential Root Proteins in Medicago sativa and Medicago truncatula in Response to Salt Stress
Abstract
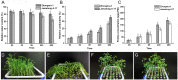
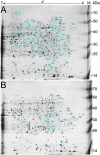
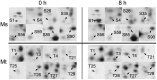
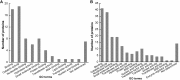
Salt stress is an important abiotic stress that causes decreased crop yields. Root growth and plant activities are affected by salt stress through the actions of specific genes that help roots adapt to adverse environmental conditions. For a more comprehensive understanding of proteins affected by salinity, we used two-dimensional gel electrophoresis and mass spectrometry to characterize the proteome-level changes associated with salt stress response in Medicago sativa cv. Zhongmu-1 and Medicago truncatula cv. Jemalong A17 roots. Our physiological and phenotypic observations indicated that Zhongmu-1 was more salt tolerant than Jemalong A17. We identified 93 and 30 proteins whose abundance was significantly affected by salt stress in Zhongmu-1 and Jemalong A17 roots, respectively. The tandem mass spectrometry analysis of the differentially accumulated proteins resulted in the identification of 60 and 26 proteins in Zhongmu-1 and Jemalong A17 roots, respectively. Function analyses indicated molecule binding and catalytic activity were the two primary functional categories. These proteins have known functions in various molecular processes, including defense against oxidative stress, metabolism, photosynthesis, protein synthesis and processing, and signal transduction. The transcript levels of four identified proteins were determined by quantitative reverse transcription polymerase chain reaction. Our results indicate that some of the identified proteins may play key roles in salt stress tolerance.
Keywords: 2-DE; Medicago; function; gene expression; protein; root; salt stress.
Figures
Similar articles
-
Quantitative proteomic analysis using iTRAQ to identify salt-responsive proteins during the germination stage of two Medicago species.Sci Rep. 2018 Jun 22;8(1):9553. doi: 10.1038/s41598-018-27935-8. Sci Rep. 2018. PMID: 29934583 Free PMC article.
-
Small RNA deep sequencing identifies novel and salt-stress-regulated microRNAs from roots of Medicago sativa and Medicago truncatula.Physiol Plant. 2015 May;154(1):13-27. doi: 10.1111/ppl.12266. Epub 2014 Oct 7. Physiol Plant. 2015. PMID: 25156209
-
Comparative Proteomic Analysis Reveals That Antioxidant System and Soluble Sugar Metabolism Contribute to Salt Tolerance in Alfalfa ( Medicago sativa L.) Leaves.J Proteome Res. 2019 Jan 4;18(1):191-203. doi: 10.1021/acs.jproteome.8b00521. Epub 2018 Oct 25. J Proteome Res. 2019. PMID: 30359026
-
Differential expression of the TFIIIA regulatory pathway in response to salt stress between Medicago truncatula genotypes.Plant Physiol. 2007 Dec;145(4):1521-32. doi: 10.1104/pp.107.106146. Epub 2007 Oct 19. Plant Physiol. 2007. PMID: 17951460 Free PMC article.
-
Medicago truncatula genotypes Jemalong A17 and R108 show contrasting variations under drought stress.Plant Physiol Biochem. 2016 Dec;109:190-198. doi: 10.1016/j.plaphy.2016.09.019. Epub 2016 Oct 1. Plant Physiol Biochem. 2016. PMID: 27721134
Cited by
-
Salt stress proteins in plants: An overview.Front Plant Sci. 2022 Dec 16;13:999058. doi: 10.3389/fpls.2022.999058. eCollection 2022. Front Plant Sci. 2022. PMID: 36589054 Free PMC article. Review.
-
Quantitative proteomic analysis using iTRAQ to identify salt-responsive proteins during the germination stage of two Medicago species.Sci Rep. 2018 Jun 22;8(1):9553. doi: 10.1038/s41598-018-27935-8. Sci Rep. 2018. PMID: 29934583 Free PMC article.
-
Physiological and Differential Proteomic Analyses of Imitation Drought Stress Response in Sorghum bicolor Root at the Seedling Stage.Int J Mol Sci. 2020 Dec 1;21(23):9174. doi: 10.3390/ijms21239174. Int J Mol Sci. 2020. PMID: 33271965 Free PMC article.
-
Salt-tolerant and -sensitive alfalfa (Medicago sativa) cultivars have large variations in defense responses to the lepidopteran insect Spodoptera litura under normal and salt stress condition.PLoS One. 2017 Jul 18;12(7):e0181589. doi: 10.1371/journal.pone.0181589. eCollection 2017. PLoS One. 2017. PMID: 28719628 Free PMC article.
-
An Insight into Abiotic Stress and Influx Tolerance Mechanisms in Plants to Cope in Saline Environments.Biology (Basel). 2022 Apr 14;11(4):597. doi: 10.3390/biology11040597. Biology (Basel). 2022. PMID: 35453796 Free PMC article. Review.
References
-
- Bates L., Waldren R., Teare I. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. 10.1007/BF00018060 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

