A transcriptional signature of Alzheimer's disease is associated with a metastable subproteome at risk for aggregation
- PMID: 27071083
- PMCID: PMC4855616
- DOI: 10.1073/pnas.1516604113
A transcriptional signature of Alzheimer's disease is associated with a metastable subproteome at risk for aggregation
Abstract
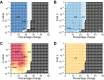
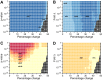
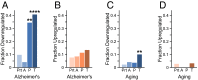
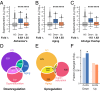
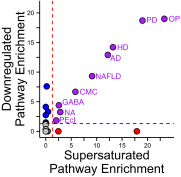
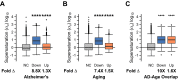
It is well-established that widespread transcriptional changes accompany the onset and progression of Alzheimer's disease. Because of the multifactorial nature of this neurodegenerative disorder and its complex relationship with aging, however, it remains unclear whether such changes are the result of nonspecific dysregulation and multisystem failure or instead are part of a coordinated response to cellular dysfunction. To address this problem in a systematic manner, we performed a meta-analysis of about 1,600 microarrays from human central nervous system tissues to identify transcriptional changes upon aging and as a result of Alzheimer's disease. Our strategy to discover a transcriptional signature of Alzheimer's disease revealed a set of down-regulated genes that encode proteins metastable to aggregation. Using this approach, we identified a small number of biochemical pathways, notably oxidative phosphorylation, enriched in proteins vulnerable to aggregation in control brains and encoded by genes down-regulated in Alzheimer's disease. These results suggest that the down-regulation of a metastable subproteome may help mitigate aberrant protein aggregation when protein homeostasis becomes compromised in Alzheimer's disease.
Keywords: amyloid formation; neurodegenerative diseases; protein aggregation; protein misfolding; protein supersaturation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Protein homeostasis of a metastable subproteome associated with Alzheimer's disease.Proc Natl Acad Sci U S A. 2017 Jul 11;114(28):E5703-E5711. doi: 10.1073/pnas.1618417114. Epub 2017 Jun 26. Proc Natl Acad Sci U S A. 2017. PMID: 28652376 Free PMC article.
-
A protein homeostasis signature in healthy brains recapitulates tissue vulnerability to Alzheimer's disease.Sci Adv. 2016 Aug 10;2(8):e1600947. doi: 10.1126/sciadv.1600947. eCollection 2016 Aug. Sci Adv. 2016. PMID: 27532054 Free PMC article.
-
The Alzheimer's disease transcriptome mimics the neuroprotective signature of IGF-1 receptor-deficient neurons.Brain. 2017 Jul 1;140(7):2012-2027. doi: 10.1093/brain/awx132. Brain. 2017. PMID: 28595357
-
β-Amyloid and the Pathomechanisms of Alzheimer's Disease: A Comprehensive View.Molecules. 2017 Oct 10;22(10):1692. doi: 10.3390/molecules22101692. Molecules. 2017. PMID: 28994715 Free PMC article. Review.
-
Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase in neurodegenerative processes and the role of low molecular weight compounds in counteracting its aggregation and nuclear translocation.Ageing Res Rev. 2018 Dec;48:21-31. doi: 10.1016/j.arr.2018.09.003. Epub 2018 Sep 22. Ageing Res Rev. 2018. PMID: 30254002 Review.
Cited by
-
The Amyloid Phenomenon and Its Significance in Biology and Medicine.Cold Spring Harb Perspect Biol. 2020 Feb 3;12(2):a033878. doi: 10.1101/cshperspect.a033878. Cold Spring Harb Perspect Biol. 2020. PMID: 30936117 Free PMC article. Review.
-
Dissecting the Molecular Mechanisms of Neurodegenerative Diseases through Network Biology.Front Aging Neurosci. 2017 May 29;9:166. doi: 10.3389/fnagi.2017.00166. eCollection 2017. Front Aging Neurosci. 2017. PMID: 28611656 Free PMC article.
-
Melatonin and Melatonergic Influence on Neuronal Transcription Factors: Implications for the Development of Novel Therapies for Neurodegenerative Disorders.Curr Neuropharmacol. 2020;18(7):563-577. doi: 10.2174/1570159X18666191230114339. Curr Neuropharmacol. 2020. PMID: 31885352 Free PMC article. Review.
-
Early Life Stress and Epigenetics in Late-onset Alzheimer's Dementia: A Systematic Review.Curr Genomics. 2018 Nov;19(7):522-602. doi: 10.2174/1389202919666171229145156. Curr Genomics. 2018. PMID: 30386171 Free PMC article. Review.
-
Deep post-GWAS analysis identifies potential risk genes and risk variants for Alzheimer's disease, providing new insights into its disease mechanisms.Sci Rep. 2021 Oct 15;11(1):20511. doi: 10.1038/s41598-021-99352-3. Sci Rep. 2021. PMID: 34654853 Free PMC article.
References
-
- Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15(6):384–396. - PubMed
-
- Querfurth HW, LaFerla FM. Alzheimer’s disease. N Engl J Med. 2010;362(4):329–344. - PubMed
-
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443(7113):780–786. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

