TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe
- PMID: 27154402
- PMCID: PMC4931564
- DOI: 10.15252/embr.201541708
TERRA promotes telomerase-mediated telomere elongation in Schizosaccharomyces pombe
Abstract
Telomerase-mediated telomere elongation provides cell populations with the ability to proliferate indefinitely. Telomerase is capable of recognizing and extending the shortest telomeres in cells; nevertheless, how this mechanism is executed remains unclear. Here, we show that, in the fission yeast Schizosaccharomyces pombe, shortened telomeres are highly transcribed into the evolutionarily conserved long noncoding RNA TERRA A fraction of TERRA produced upon telomere shortening is polyadenylated and largely devoid of telomeric repeats, and furthermore, telomerase physically interacts with this polyadenylated TERRA in vivo We also show that experimentally enhanced transcription of a manipulated telomere promotes its association with telomerase and concomitant elongation. Our data represent the first direct evidence that TERRA stimulates telomerase recruitment and activity at chromosome ends in an organism with human-like telomeres.
Keywords: TERRA; fission yeast; telomerase; telomere length regulation; transcription.
© 2016 The Authors.
Figures

Schematic representation of the S. pombe telomeric transcriptome. Telomeric repeats are shown in black and subtelomeric sequences in gray. Oligonucleotides used for RT–PCR and for 3′ end RACE are indicated.
Growth curves of the indicated strains shown as population doublings (pds) on the y‐axis and hours in liquid culture on the x‐axis. Before inoculation in liquid YES medium, single cells underwent approximately 25 pds to produce visible colonies on solid medium. For each strain, averages and SDs (error bars) of three biological replicates are shown. Arrows indicate times of harvesting and corresponding pds for experiments are shown in (C) and (D).
Telo‐PCR analysis of telomere length in wt (ter1+ and est1+) and two independent clones (a and b) deleted for ter1+ or est1+ grown for ˜25 + 10 (˜35), ˜25 + 14 (˜39), and ˜25 + 18 (˜43) pds in YES medium. PCRs were performed with the subtelomeric oligonucleotide oF5, located 620 bp upstream of the first telomeric repeat, and the oligo(dG)‐containing oligonucleotide odG18. Marker (m) molecular weights are on the left in kilobases. tels: telomere‐containing Telo‐PCR products.
qRT–PCR analysis of G‐rich TERRA, polyA+ TERRA, and ARRET/αARRET levels in cells as in (B). Values are normalized to ACT1 mRNA and expressed as fold increase over control wt strains (ter1+ and est1+) grown in parallel. Bars and error bars are averages and SD from 4 (ter1Δ clones) or 3 (est1Δ clones) independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (relative to wt; two‐tailed Student's t‐test).
ChIP experiments with anti‐RNA polymerase II (RPII) antibodies and extracts from ter1+ and ter1Δ cells grown for ˜43 pds. Quantitative PCRs were performed using oligonucleotides oF1 + oR1 (subtel) or oligonucleotides amplifying a region from the actively transcribed act1+ locus. Values are expressed as fold increase over ter1+. Bars and error bars are averages and SD from three independent experiments. *P < 0.05 (relative to ter1+; two‐tailed Student's t‐test). The Western blot to the right shows no difference in RNPII levels between ter1+ and ter1Δ cells used in ChIP experiments. Ponceau S staining serves as loading control.
3′ RACE analysis of polyA+ TERRA in ter1Δ cells grown for ˜43 pds. The sequence at the top is the genomic reference sequence (gen) taken from the pNSU70 plasmid (nucleotide positions are indicated). The sequences below derive from all positive clones isolated. Sequences boxed in blue, green, and yellow correspond to the oligonucleotide used for RACE (o3′RACE), subtelomeric DNA, and telomeric repeats, respectively. Genomic adenines corresponding to putative RNA cleavage sites (CS) are highlighted.
Western blot analysis of cellular fractions from ter1+ and ter1Δ cells grown for ˜43 pds. Total proteins (tot) and equivalents of soluble (sol) and insoluble (ins) proteins were loaded. Actin serves as a control for soluble proteins and H3K9ac for chromatin‐bound proteins.
qRT–PCR analysis of total TERRA and polyA+ TERRA in cellular fractions as in (G). Values are expressed after normalization to total cellular extracts. Bars and error bars are averages and SD from 3 independent experiments.

RIP experiments were performed using anti‐myc antibodies and extracts from Trt1‐myc‐expressing strains followed by qRT–PCR to detect G‐rich TERRA, polyA+ TERRA, ARRET, TER1, and ACT1. Values represent fractions of input RNA detected in immunoprecipitated material expressed as fold increase over untagged (unt) wt strain. Bars and error bars are averages and SD from three independent experiments. *P < 0.05 (relative to unt; two‐tailed Student's t‐test).
RIP experiments as in (A) where, prior to RNA extraction, washed beads were resuspended in DNaseI buffer (DNbuff) and incubated for 1 hour at 30°C in the presence or absence of DNaseI. Values represent fractions of input polyA+ TERRA detected in immunoprecipitated material expressed as fold increase over an untagged (unt) wt strain. Bars and error bars are averages and SD from three independent experiments. *P < 0.05 (relative to unt; two‐tailed Student's t‐test).
RIP experiments were performed using anti‐myc antibodies and extracts from Trt1‐myc‐expressing strains (either ter1+ or ter1Δ) cultured in YES medium for ˜43 pds. Values represent polyA+ TERRA detected in immunoprecipitated material expressed as a fraction of the input (graph on the left) or after normalization to ACT1 RNA in the corresponding input (on the right), and expressed as fold increase over an untagged wt strain (unt ter1+). Bars and error bars are averages and SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (relative to unt ter1+; two‐tailed Student's t‐test).
Western blot analysis of Trt1‐myc in cells used in (C). Ponceau S staining serves as a loading control.

Schematic representation of tiTELs. An nmt1 promoter (Pnmt1) was inserted upstream of the natural TERRA TSS. Telomeric repeats are in black and the subtelomeric tract in gray. A cassette conferring resistance to nourseothricin (nat r) was used to select for positively transformed cells. The inserted cassette generates a novel HindIII restriction site, while the endogenous ApaI restriction site close to the telomeric tract is disrupted upon integration. Probes and oligonucleotides used for Northern blot, Southern blot, and RT–PCR analyses are indicated. The sketch is not to scale.
Genomic DNA from wt cells (CAF13) and cells carrying two tiTELs (CAF110) maintained in YES medium was digested with HindIII and Southern blot‐hybridized consecutively with the indicated probes. RET refers to telomeric sequences (tiTELs or natural telomeres, nTELs) retained in the upper part of the gel; REL refers to telomeric sequences released in the lower part of the gel. The black arrow points to the natural nmt1+ sequence on chromosome III. Marker molecular weights are on the left in kilobases.
5′ RACE analysis of the tiTERRA transcription start site using total RNA from cells carrying two tiTELs (CAF110) maintained in YES medium. Nucleotides boxed in blue overlap with the 5′ UTR of the nmt1+ transcript, while the ones in green correspond to subtelomeric sequences. tiTERRA and TERRA transcription start sites (TSS) are indicated.
qRT–PCR analysis of tiTERRA in cells carrying no tiTEL (CAF13), one tiTEL (CAF545), or two tiTELs (CAF110) cultured in EMM in the presence or absence of thiamine (THI; repressed and induced condition, respectively) for 24 h. RNA was reverse‐transcribed with oC, and cDNA was amplified with oF3 and oR3 oligonucleotides to detect only tiTERRA (graph on the left) or with oF1 and oR1 oligonucleotides to detect simultaneously tiTERRA and natural TERRA (on the right). Values are normalized to ACT1 mRNA and expressed as fold increase over CAF545 in uninduced conditions (THI+). Bars and error bars are averages and SD from 3 independent experiments. *P < 0.05, **P < 0.01 (relative to CAF545 THI+; two‐tailed Student's t‐test).
Northern blot analysis of total RNA from wt cells and cells carrying two tiTELs (CAF110) treated with combinations of THI and trichostatin A (TSA) for 24 h. Two identical membranes were hybridized in parallel with oligonucleotides corresponding to C‐rich (oC) or G‐rich (oG) telomeric repeats. To confirm similar hybridization efficiencies of the two oligonucleotides, Northern blot membranes were simultaneously hybridized and exposed along with blots of digested genomic DNA (upper right insets). After signal detection, RNA membranes were stripped and rehybridized with U6 probes to assure equal loading. Marker molecular weights are on the left in kilobases.

Pulsed‐field gel electrophoresis (PFGE) analysis of Pnmt1 insertion (ins) in tiTEL strains. Genomic DNA from wt cells (CAF13) and cells carrying one tiTEL (CF545) or two tiTELs (CAF110) maintained in YES medium was digested with NotI, separated by PFGE, and hybridized sequentially using nmt1 and telomeric probes. The positions and nomenclature of the four NotI terminal restriction fragments entering the gel are indicated on the right. Genomic organization of tiTEL strains is sketched on the right.
Genomic DNA was extracted from wt (CAF13) cells and cells carrying two tiTELs (CAF110) cultured for 24 h in EMM with THI and TSA as indicated. DNA was digested with ApaI and hybridized sequentially using nmt1 and telomeric probes. The asterisk indicates a band corresponding the endogenous nmt1+. TiTELs and natural telomeres (nTELs) are indicated. REL refers to telomeric sequences released in the lower part of the gel. Marker molecular weights are on the left in kilobases.
Control RT–PCR experiments showing the specificity of oF3 + oR3 oligonucleotides. Nucleic acids were prepared from wt cells (CAF13) and cells carrying one tiTEL (CAF545) cultured for 24 h in EMM without THI. gD indicates genomic DNA, RT indicates whether or not samples were treated with reverse transcriptase. ACT1 PCR was performed to assure presence of genomic DNA and cDNA templates. Marker (m) molecular weights are on the left in base pairs.
Telo‐PCR analysis of tiTELs using genomic DNA from cells carrying one tiTEL (CAF545, on the left) or two tiTELs (CAF110, on the right) cultured for 24 h in EMM with THI and TSA as indicated. Marker (m) molecular weights are shown in base pairs.
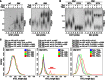
Telomere restriction fragment analysis of genomic DNA from wt cells and cells carrying two tiTELs (CAF110) grown for 24 h in EMM containing THI and TSA as indicated. DNA was digested with HindIII and hybridized first with a nmt1 probe and successively with a telomeric probe. Marker molecular weights are on the left in kilobases. Numbers at the bottom indicate gel lanes.
Genomic DNA from cells carrying two tiTELs and deleted for trt1+ (CAF113) was analyzed as in (A). Marker molecular weights are on the left in kilobases. Numbers at the bottom indicate gel lanes.
ter1+ cells carrying two tiTELs (CAF110, left panel) or trt1Δ cells carrying one tiTEL (MKSP2104, central panel) were cultured in EMM in the presence of THI for 24 h (THI+) or in the absence of THI for 24, 48, or 72 h. In the right panel, cells carrying two tiTELs (CAF110) were treated with TSA for 24 h in the presence or absence of THI. TiTEL telomeric sequences were amplified from genomic DNA and sequenced using a PacBio platform. The statistical analysis for each set of conditions is displayed above each panel. P‐values for each comparison were derived from the Student's t‐test between each condition and the THI+ sample using Welch's correction when appropriate. The new population of longer tiTELs appearing in trt1Δ cells upon prolonged transcription induction is indicated in red (center panel).
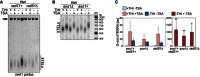
Genomic DNA was extracted from cells carrying one tiTEL either rad51+ (CAF545) or rad51Δ (CAF550) cultured for 24 h in EMM with THI and TSA as indicated. DNA was digested with HindIII and Southern blot‐hybridized with nmt1 probes. The black arrow points to the nmt1+ sequence on chromosome III.
Telo‐PCR analysis of tiTELs in cells carrying one tiTEL either exo1+ (CAF545) or exo1Δ (CAF655) cultured for 24 h in EMM with THI and TSA as indicated. Marker (m) molecular weights are on the right in base pairs.
qRT–PCR analysis of total tiTERRA levels in cells as in (A) and (B). Values are expressed as fold increase over uninduced exo1+ rad51+ strain (CAF545). Bars and error bars are averages and SD from 3 independent experiments. *P < 0.05, **P < 0.01 (relative to uninduced parental; two‐tailed Student's t‐test).

Genomic DNA was isolated from wt cells (CAF13) and cells carrying two tiTELs (CAF110) cultured for 7 days in EMM with or without THI. DNA was digested and hybridized as indicated. Marker molecular weights are on the left in kilobases.
Sketch showing the positions of oligonucleotides used for Telo‐PCR amplification and sequencing of tiTELs and natural telomeres from the same cell. Telo‐PCRs were performed using oF4 or oF5 in combination with the G‐rich oligonucleotide odG18. Sequencing was performed with oF6.
Examples of Telo‐PCR products using genomic DNA from tiTEL cells as in (A) and amplified using the indicated oligonucleotides. Marker (m) molecular weights are on the left in base pairs.
Telomere PCR products corresponding to tiTELs (on the left) or nTELs (on the right) from tiTEL cells as in (A) were cloned and sequenced. Blue bars represent individual telomeres plotted against their length in base pairs (y‐axis). Average telomere length of the sequenced population (av) and standard deviations (SD) are indicated along with P‐values (−THI vs. +THI) calculated with the two‐tailed Student's t‐test.
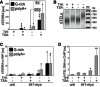
qRT–PCR analysis of G‐rich and polyA+ tiTERRA in cells carrying one tiTEL and expressing Trt1‐myc (CAF610) cultured in EMM with the indicated combinations of THI and TSA for 24 h. Values are normalized to ACT1 mRNA and expressed as fold increase over THI+ TSA− samples. Bars and error bars are averages and SD from 4 independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (relative to THI+ TSA−; two‐tailed Student's t‐test).
Telo‐PCR analysis of tiTELs using genomic DNA extracted from cells as in (A). Marker (m) molecular weights are on the right in base pairs.
RIP experiments performed using anti‐myc antibodies and extracts from cells as in (A) followed by qRT–PCR analysis of G‐rich and polyA+ tiTERRA. Values correspond to fraction of input RNA expressed as fold increase over an untagged (unt) control tiTEL strain (CAF545). Bars and error bars are averages and SD from three independent experiments. *P < 0.05, **P < 0.01 (relative to unt; two‐tailed Student's t‐test).
ChIP experiments performed using anti‐myc antibodies and extracts from cells as in (A) followed by qPCR using tiTEL‐specific oligonucleotides. Values correspond to fraction of input DNA expressed as fold increase over THI+ TSA−. Unt: untagged strain (CAF545). Bars and error bars are averages and SD from at least three independent experiments. *P < 0.05, **P < 0.01 (relative to THI+ TSA−; two‐tailed Student's t‐test).
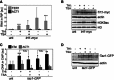
RIP experiments performed using anti‐myc antibodies and extracts from cells carrying one tiTEL and expressing Trt1‐myc (CAF610) cultured for 24 h in EMM with THI and TSA as indicated. TER1 (positive control) and ACT1 (negative control) in input and immunoprecipitated material were quantified by qRT–PCR. Values correspond to fraction of input RNA expressed as fold increase over an untagged (unt) control tiTEL strain (CAF545). Bars and error bars are averages and SD from three independent experiments. *P < 0.05, ***P < 0.001 (relative to unt; two‐tailed Student's t‐test).
Western blot analysis of Trt1‐myc and histone H3 acetylated at lysine 9 (H3K9ac) levels in cells as in (A). Actin and total H3 serve as loading controls. The accumulation of H3K9ac in TSA+ cells confirms the effectiveness of TSA. The two upper panels and the two lower panels are from two independent membranes.
ChIP experiments were performed using anti‐GFP antibodies and extracts from cells carrying one tiTEL and expressing Taz1‐GFP (MKSP1781) cultured for 24 h in EMM with THI and TSA as indicated followed by tiTEL qPCR. ACT1 qPCRs were performed to control for specificity. Values correspond to fraction of input DNA expressed as fold increase over an untagged control tiTEL strain (CAF545). Bars and error bars are averages and SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 (relative to unt; two‐tailed Student's t‐test).
Western blot analysis of Taz1‐GFP protein levels in cells as in (C). Actin serves as a loading control.
Comment in
-
TERRA Incognita at chromosome ends.EMBO Rep. 2016 Jul;17(7):933-4. doi: 10.15252/embr.201642583. Epub 2016 Jun 3. EMBO Rep. 2016. PMID: 27259461 Free PMC article.
Similar articles
-
RNA-DNA Hybrids Support Recombination-Based Telomere Maintenance in Fission Yeast.Genetics. 2019 Oct;213(2):431-447. doi: 10.1534/genetics.119.302606. Epub 2019 Aug 12. Genetics. 2019. PMID: 31405990 Free PMC article.
-
TERRA transcripts localize at long telomeres to regulate telomerase access to chromosome ends.Sci Adv. 2024 Jun 14;10(24):eadk4387. doi: 10.1126/sciadv.adk4387. Epub 2024 Jun 12. Sci Adv. 2024. PMID: 38865460 Free PMC article.
-
Tpz1TPP1 prevents telomerase activation and protects telomeres by modulating the Stn1-Ten1 complex in fission yeast.Commun Biol. 2019 Aug 7;2:297. doi: 10.1038/s42003-019-0546-8. eCollection 2019. Commun Biol. 2019. PMID: 31396577 Free PMC article.
-
The makings of TERRA R-loops at chromosome ends.Cell Cycle. 2021 Sep;20(18):1745-1759. doi: 10.1080/15384101.2021.1962638. Epub 2021 Aug 25. Cell Cycle. 2021. PMID: 34432566 Free PMC article. Review.
-
Protection and replication of telomeres in fission yeast.Biochem Cell Biol. 2009 Oct;87(5):747-58. doi: 10.1139/O09-037. Biochem Cell Biol. 2009. PMID: 19898524 Free PMC article. Review.
Cited by
-
Dynamics of telomeric repeat-containing RNA expression in early embryonic cleavage stages with regards to maternal age.Aging (Albany NY). 2020 Aug 29;12(16):15906-15917. doi: 10.18632/aging.103922. Epub 2020 Aug 29. Aging (Albany NY). 2020. PMID: 32860669 Free PMC article.
-
TERRA expression is regulated by the telomere-binding proteins POT-1 and POT-2 in Caenorhabditis elegans.Nucleic Acids Res. 2023 Oct 27;51(19):10681-10699. doi: 10.1093/nar/gkad742. Nucleic Acids Res. 2023. PMID: 37713629 Free PMC article.
-
From "Cellular" RNA to "Smart" RNA: Multiple Roles of RNA in Genome Stability and Beyond.Chem Rev. 2018 Apr 25;118(8):4365-4403. doi: 10.1021/acs.chemrev.7b00487. Epub 2018 Mar 30. Chem Rev. 2018. PMID: 29600857 Free PMC article. Review.
-
Paf1 and Ctr9, core components of the PAF1 complex, maintain low levels of telomeric repeat containing RNA.Nucleic Acids Res. 2018 Jan 25;46(2):621-634. doi: 10.1093/nar/gkx1131. Nucleic Acids Res. 2018. PMID: 29145644 Free PMC article.
-
Telomere Fragility and MiDAS: Managing the Gaps at the End of the Road.Genes (Basel). 2023 Jan 29;14(2):348. doi: 10.3390/genes14020348. Genes (Basel). 2023. PMID: 36833275 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

