PACRG, a protein linked to ciliary motility, mediates cellular signaling
- PMID: 27193298
- PMCID: PMC4927285
- DOI: 10.1091/mbc.E15-07-0490
PACRG, a protein linked to ciliary motility, mediates cellular signaling
Abstract
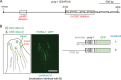
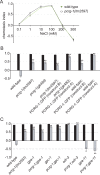
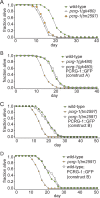
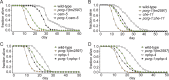
Cilia are microtubule-based organelles that project from nearly all mammalian cell types. Motile cilia generate fluid flow, whereas nonmotile (primary) cilia are required for sensory physiology and modulate various signal transduction pathways. Here we investigate the nonmotile ciliary signaling roles of parkin coregulated gene (PACRG), a protein linked to ciliary motility. PACRG is associated with the protofilament ribbon, a structure believed to dictate the regular arrangement of motility-associated ciliary components. Roles for protofilament ribbon-associated proteins in nonmotile cilia and cellular signaling have not been investigated. We show that PACRG localizes to a small subset of nonmotile cilia in Caenorhabditis elegans, suggesting an evolutionary adaptation for mediating specific sensory/signaling functions. We find that it influences a learning behavior known as gustatory plasticity, in which it is functionally coupled to heterotrimeric G-protein signaling. We also demonstrate that PACRG promotes longevity in C. elegans by acting upstream of the lifespan-promoting FOXO transcription factor DAF-16 and likely upstream of insulin/IGF signaling. Our findings establish previously unrecognized sensory/signaling functions for PACRG and point to a role for this protein in promoting longevity. Furthermore, our work suggests additional ciliary motility-signaling connections, since EFHC1 (EF-hand containing 1), a potential PACRG interaction partner similarly associated with the protofilament ribbon and ciliary motility, also positively regulates lifespan.
© 2016 Loucks, Bialas, et al. This article is distributed by The American Society for Cell Biology under license from the author(s). Two months after publication it is available to the public under an Attribution–Noncommercial–Share Alike 3.0 Unported Creative Commons License (http://creativecommons.org/licenses/by-nc-sa/3.0).
Figures




Similar articles
-
Microtubule binding protein PACRG plays a role in regulating specific ciliary dyneins during microtubule sliding.Cytoskeleton (Hoboken). 2016 Dec;73(12):703-711. doi: 10.1002/cm.21340. Epub 2016 Nov 8. Cytoskeleton (Hoboken). 2016. PMID: 27770595 Free PMC article.
-
PACRG and FAP20 form the inner junction of axonemal doublet microtubules and regulate ciliary motility.Mol Biol Cell. 2019 Jul 15;30(15):1805-1816. doi: 10.1091/mbc.E19-01-0063. Epub 2019 May 22. Mol Biol Cell. 2019. PMID: 31116684 Free PMC article.
-
CDKL kinase regulates the length of the ciliary proximal segment.Curr Biol. 2021 Jun 7;31(11):2359-2373.e7. doi: 10.1016/j.cub.2021.03.068. Epub 2021 Apr 14. Curr Biol. 2021. PMID: 33857430
-
DAF-16/FoxO in Caenorhabditis elegans and Its Role in Metabolic Remodeling.Cells. 2020 Jan 2;9(1):109. doi: 10.3390/cells9010109. Cells. 2020. PMID: 31906434 Free PMC article. Review.
-
DAF-16: FOXO in the Context of C. elegans.Curr Top Dev Biol. 2018;127:1-21. doi: 10.1016/bs.ctdb.2017.11.007. Epub 2018 Feb 2. Curr Top Dev Biol. 2018. PMID: 29433733 Review.
Cited by
-
Functional Expression, Purification and Identification of Interaction Partners of PACRG.Molecules. 2021 Apr 16;26(8):2308. doi: 10.3390/molecules26082308. Molecules. 2021. PMID: 33923444 Free PMC article.
-
Rare copy number variants analysis identifies novel candidate genes in heterotaxy syndrome patients with congenital heart defects.Genome Med. 2018 May 30;10(1):40. doi: 10.1186/s13073-018-0549-y. Genome Med. 2018. PMID: 29843777 Free PMC article.
-
Crystal structure of human PACRG in complex with MEIG1 reveals roles in axoneme formation and tubulin binding.Structure. 2021 Jun 3;29(6):572-586.e6. doi: 10.1016/j.str.2021.01.001. Epub 2021 Feb 1. Structure. 2021. PMID: 33529594 Free PMC article.
-
Doublet microtubule inner junction protein FAP20 recruits tubulin to the microtubule lattice.Structure. 2023 Dec 7;31(12):1535-1544.e4. doi: 10.1016/j.str.2023.09.010. Epub 2023 Oct 9. Structure. 2023. PMID: 37816351 Free PMC article.
-
Modulation of inner junction proteins contributes to axoneme differentiation.Proc Natl Acad Sci U S A. 2023 Jul 25;120(30):e2303955120. doi: 10.1073/pnas.2303955120. Epub 2023 Jul 18. Proc Natl Acad Sci U S A. 2023. PMID: 37463209 Free PMC article.
References
-
- Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2004;41:45–55. - PubMed
-
- Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. - PubMed
-
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

