GpDSR7, a Novel E3 Ubiquitin Ligase Gene in Grimmia pilifera Is Involved in Tolerance to Drought Stress in Arabidopsis
- PMID: 27228205
- PMCID: PMC4882056
- DOI: 10.1371/journal.pone.0155455
GpDSR7, a Novel E3 Ubiquitin Ligase Gene in Grimmia pilifera Is Involved in Tolerance to Drought Stress in Arabidopsis
Abstract
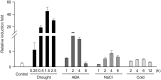
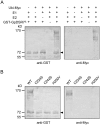
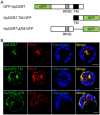
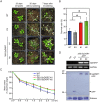
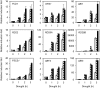
The growth and development of plants under drought stress depends mainly on the expression levels of various genes and modification of proteins. To clarify the molecular mechanism of drought-tolerance of plants, suppression subtractive hybridisation cDNA libraries were screened to identify drought-stress-responsive unigenes in Grimmia pilifera, and a novel E3 ubiquitin ligase gene, GpDSR7, was identified among the 240 responsive unigenes. GpDSR7 expression was induced by various abiotic stresses, particularly by drought. GpDSR7 displayed E3 ubiquitin ligase activity in vitro and was exclusively localised on the ER membrane in Arabidopsis mesophyll protoplasts. GpDSR7-overexpressing transgenic Arabidopsis plants showed a high water content and survival ratio under drought stress. Moreover, the expression levels of some marker genes involved in drought stress were higher in the transgenic plants than in wild-type plants. These results suggest that GpDSR7, an E3 ubiquitin ligase, is involved in tolerance to drought stress at the protein modification level.
Conflict of interest statement
Figures

Similar articles
-
Genome-Wide Identification of Soybean U-Box E3 Ubiquitin Ligases and Roles of GmPUB8 in Negative Regulation of Drought Stress Response in Arabidopsis.Plant Cell Physiol. 2016 Jun;57(6):1189-209. doi: 10.1093/pcp/pcw068. Epub 2016 Apr 6. Plant Cell Physiol. 2016. PMID: 27057003
-
Heterologous expression of the gourd E3 ubiquitin ligase gene LsRZF1 compromises the drought stress tolerance in Arabidopsis thaliana.Plant Physiol Biochem. 2014 Apr;77:7-14. doi: 10.1016/j.plaphy.2014.01.010. Epub 2014 Jan 25. Plant Physiol Biochem. 2014. PMID: 24525351
-
RING E3 ubiquitin ligase TaSADR1 negatively regulates drought resistance in transgenic Arabidopsis.Plant Physiol Biochem. 2022 Jan 1;170:255-265. doi: 10.1016/j.plaphy.2021.12.004. Epub 2021 Dec 8. Plant Physiol Biochem. 2022. PMID: 34922142
-
RING E3 ligases: key regulatory elements are involved in abiotic stress responses in plants.BMB Rep. 2017 Aug;50(8):393-400. doi: 10.5483/bmbrep.2017.50.8.128. BMB Rep. 2017. PMID: 28712388 Free PMC article. Review.
-
Plant E3 Ligases and Their Role in Abiotic Stress Response.Cells. 2022 Mar 4;11(5):890. doi: 10.3390/cells11050890. Cells. 2022. PMID: 35269512 Free PMC article. Review.
Cited by
-
Endoplasmic reticulum-related E3 ubiquitin ligases: Key regulators of plant growth and stress responses.Plant Commun. 2021 Apr 16;2(3):100186. doi: 10.1016/j.xplc.2021.100186. eCollection 2021 May 10. Plant Commun. 2021. PMID: 34027397 Free PMC article. Review.
-
Identification of C3H2C3-type RING E3 ubiquitin ligase in grapevine and characterization of drought resistance function of VyRCHC114.BMC Plant Biol. 2021 Sep 17;21(1):422. doi: 10.1186/s12870-021-03162-8. BMC Plant Biol. 2021. PMID: 34535070 Free PMC article.
-
Overexpression of E3 Ubiquitin Ligase Gene AdBiL Contributes to Resistance against Chilling Stress and Leaf Mold Disease in Tomato.Front Plant Sci. 2017 Jun 30;8:1109. doi: 10.3389/fpls.2017.01109. eCollection 2017. Front Plant Sci. 2017. PMID: 28713400 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

