Long Terminal Repeats: From Parasitic Elements to Building Blocks of the Transcriptional Regulatory Repertoire
- PMID: 27259207
- PMCID: PMC4910160
- DOI: 10.1016/j.molcel.2016.03.029
Long Terminal Repeats: From Parasitic Elements to Building Blocks of the Transcriptional Regulatory Repertoire
Abstract
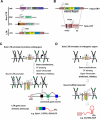
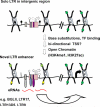
The life cycle of endogenous retroviruses (ERVs), also called long terminal repeat (LTR) retrotransposons, begins with transcription by RNA polymerase II followed by reverse transcription and re-integration into the host genome. While most ERVs are relics of ancient integration events, "young" proviruses competent for retrotransposition-found in many mammals, but not humans-represent an ongoing threat to host fitness. As a consequence, several restriction pathways have evolved to suppress their activity at both transcriptional and post-transcriptional stages of the viral life cycle. Nevertheless, accumulating evidence has revealed that LTR sequences derived from distantly related ERVs have been exapted as regulatory sequences for many host genes in a wide range of cell types throughout mammalian evolution. Here, we focus on emerging themes from recent studies cataloging the diversity of ERV LTRs acting as important transcriptional regulatory elements in mammals and explore the molecular features that likely account for LTR exaptation in developmental and tissue-specific gene regulation.
Published by Elsevier Inc.
Figures
Similar articles
-
Existence of Two Distinct Infectious Endogenous Retroviruses in Domestic Cats and Their Different Strategies for Adaptation to Transcriptional Regulation.J Virol. 2016 Sep 29;90(20):9029-45. doi: 10.1128/JVI.00716-16. Print 2016 Oct 15. J Virol. 2016. PMID: 27466428 Free PMC article.
-
Mammalian Endogenous Retroviruses.Microbiol Spectr. 2015 Feb;3(1):MDNA3-0009-2014. doi: 10.1128/microbiolspec.MDNA3-0009-2014. Microbiol Spectr. 2015. PMID: 26104559 Review.
-
Endogenous retroviral LTRs as promoters for human genes: a critical assessment.Gene. 2009 Dec 15;448(2):105-14. doi: 10.1016/j.gene.2009.06.020. Epub 2009 Jul 3. Gene. 2009. PMID: 19577618 Review.
-
Endogenous retrovirus long terminal repeats as ready-to-use mobile promoters: the case of primate beta3GAL-T5.Gene. 2005 Dec 30;364:2-12. doi: 10.1016/j.gene.2005.05.045. Epub 2005 Aug 22. Gene. 2005. PMID: 16112824
-
Role of human endogenous retroviral long terminal repeats (LTRs) in maintaining the integrity of the human germ line.Viruses. 2011 Jun;3(6):901-5. doi: 10.3390/v3060901. Epub 2011 Jun 21. Viruses. 2011. PMID: 21994760 Free PMC article.
Cited by
-
Endogenous Retroviruses Walk a Fine Line between Priming and Silencing.Viruses. 2020 Jul 23;12(8):792. doi: 10.3390/v12080792. Viruses. 2020. PMID: 32718022 Free PMC article. Review.
-
CRISPR-Mediated Activation of Transposable Elements in Embryonic Stem Cells.Methods Mol Biol. 2022;2509:171-194. doi: 10.1007/978-1-0716-2380-0_11. Methods Mol Biol. 2022. PMID: 35796964
-
Transposable Element Expression in Acute Myeloid Leukemia Transcriptome and Prognosis.Sci Rep. 2018 Nov 6;8(1):16449. doi: 10.1038/s41598-018-34189-x. Sci Rep. 2018. PMID: 30401833 Free PMC article.
-
Transposable Elements: Major Players in Shaping Genomic and Evolutionary Patterns.Cells. 2022 Mar 19;11(6):1048. doi: 10.3390/cells11061048. Cells. 2022. PMID: 35326499 Free PMC article. Review.
-
The Long Terminal Repeats of ERV6 Are Activated in Pre-Implantation Embryos of Cynomolgus Monkey.Cells. 2021 Oct 9;10(10):2710. doi: 10.3390/cells10102710. Cells. 2021. PMID: 34685690 Free PMC article.
References
-
- Britten RJ, Davidson EH. Gene regulation for higher cells: a theory. Science. 1969;165:349–357. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

