Two Escape Mechanisms of Influenza A Virus to a Broadly Neutralizing Stalk-Binding Antibody
- PMID: 27351973
- PMCID: PMC4924800
- DOI: 10.1371/journal.ppat.1005702
Two Escape Mechanisms of Influenza A Virus to a Broadly Neutralizing Stalk-Binding Antibody
Abstract
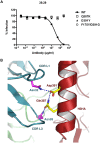
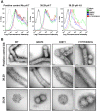
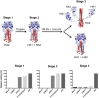
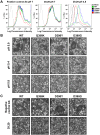
Broadly neutralizing antibodies _targeting the stalk region of influenza A virus (IAV) hemagglutinin (HA) are effective in blocking virus infection both in vitro and in vivo. The highly conserved epitopes recognized by these antibodies are critical for the membrane fusion function of HA and therefore less likely to be permissive for virus mutational escape. Here we report three resistant viruses of the A/Perth/16/2009 strain that were selected in the presence of a broadly neutralizing stalk-binding antibody. The three resistant viruses harbor three different mutations in the HA stalk: (1) Gln387Lys; (2) Asp391Tyr; (3) Asp391Gly. The Gln387Lys mutation completely abolishes binding of the antibody to the HA stalk epitope. The other two mutations, Asp391Tyr and Asp391Gly, do not affect antibody binding at neutral pH and only slightly reduce binding at low pH. Interestingly, they enhance the fusion ability of the HA, representing a novel mechanism that allows productive membrane fusion even in the presence of antibody and hence virus escape from antibody neutralization. Therefore, these mutations illustrate two different resistance mechanisms used by IAV to escape broadly neutralizing stalk-binding antibodies. Compared to the wild type virus, the resistant viruses release fewer progeny viral particles during replication and are more sensitive to Tamiflu, suggesting reduced viral fitness.
Conflict of interest statement
I have read the journal's policy and the authors of this manuscript have the following competing interests: NC, MR, EL, AF, PL, and MWT are employees of Genentech; LRS is an employee of Achaogen. This does not alter our adherence to all PLoS Pathogens policies on sharing data and materials.
Figures
Similar articles
-
A Perspective on the Structural and Functional Constraints for Immune Evasion: Insights from Influenza Virus.J Mol Biol. 2017 Aug 18;429(17):2694-2709. doi: 10.1016/j.jmb.2017.06.015. Epub 2017 Jun 23. J Mol Biol. 2017. PMID: 28648617 Free PMC article. Review.
-
Potential Role of Nonneutralizing IgA Antibodies in Cross-Protective Immunity against Influenza A Viruses of Multiple Hemagglutinin Subtypes.J Virol. 2020 Jun 1;94(12):e00408-20. doi: 10.1128/JVI.00408-20. Print 2020 Jun 1. J Virol. 2020. PMID: 32269119 Free PMC article.
-
Mutations in Influenza A Virus Neuraminidase and Hemagglutinin Confer Resistance against a Broadly Neutralizing Hemagglutinin Stem Antibody.J Virol. 2019 Jan 4;93(2):e01639-18. doi: 10.1128/JVI.01639-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30381484 Free PMC article.
-
Conformational Stability of the Hemagglutinin of H5N1 Influenza A Viruses Influences Susceptibility to Broadly Neutralizing Stem Antibodies.J Virol. 2018 May 29;92(12):e00247-18. doi: 10.1128/JVI.00247-18. Print 2018 Jun 15. J Virol. 2018. PMID: 29593038 Free PMC article.
-
Influenza Hemagglutinin Structures and Antibody Recognition.Cold Spring Harb Perspect Med. 2020 Aug 3;10(8):a038778. doi: 10.1101/cshperspect.a038778. Cold Spring Harb Perspect Med. 2020. PMID: 31871236 Free PMC article. Review.
Cited by
-
Characteristics and evolution of hemagglutinin and neuraminidase genes of Influenza A(H3N2) viruses in Thailand during 2015 to 2018.PeerJ. 2024 Jun 3;12:e17523. doi: 10.7717/peerj.17523. eCollection 2024. PeerJ. 2024. PMID: 38846750 Free PMC article.
-
Phase 2 Randomized Trial of the Safety and Efficacy of MHAA4549A, a Broadly Neutralizing Monoclonal Antibody, in a Human Influenza A Virus Challenge Model.Antimicrob Agents Chemother. 2017 Oct 24;61(11):e01154-17. doi: 10.1128/AAC.01154-17. Print 2017 Nov. Antimicrob Agents Chemother. 2017. PMID: 28807912 Free PMC article. Clinical Trial.
-
A Perspective on the Structural and Functional Constraints for Immune Evasion: Insights from Influenza Virus.J Mol Biol. 2017 Aug 18;429(17):2694-2709. doi: 10.1016/j.jmb.2017.06.015. Epub 2017 Jun 23. J Mol Biol. 2017. PMID: 28648617 Free PMC article. Review.
-
Dual neutralization of influenza virus hemagglutinin and neuraminidase by a bispecific antibody leads to improved antiviral activity.Mol Ther. 2024 Oct 2;32(10):3712-3728. doi: 10.1016/j.ymthe.2024.07.023. Epub 2024 Jul 31. Mol Ther. 2024. PMID: 39086132 Free PMC article.
-
How single mutations affect viral escape from broad and narrow antibodies to H1 influenza hemagglutinin.Nat Commun. 2018 Apr 11;9(1):1386. doi: 10.1038/s41467-018-03665-3. Nat Commun. 2018. PMID: 29643370 Free PMC article.
References
-
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, et al. (2003) Mortality associated with influenza and respiratory syncytial virus in the United States. JAMA 289: 179–186. - PubMed
-
- Thompson WW, Shay DK, Weintraub E, Brammer L, Bridges CB, et al. (2004) Influenza-associated hospitalizations in the United States. JAMA 292: 1333–1340. - PubMed
-
- Murray CJ, Lopez AD, Chin B, Feehan D, Hill KH (2006) Estimation of potential global pandemic influenza mortality on the basis of vital registry data from the 1918–20 pandemic: a quantitative analysis. Lancet 368: 2211–2218. - PubMed
-
- Pebody R, Warburton F, Ellis J, Andrews N, Thompson C, et al. (2015) Low effectiveness of seasonal influenza vaccine in preventing laboratory-confirmed influenza in primary care in the United Kingdom: 2014/15 mid-season results. Euro Surveill 20. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

