Comparative genomics of the human, macaque and mouse major histocompatibility complex
- PMID: 27395034
- PMCID: PMC5214800
- DOI: 10.1111/imm.12624
Comparative genomics of the human, macaque and mouse major histocompatibility complex
Abstract
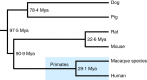
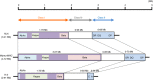
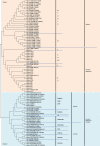
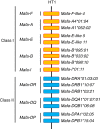
The MHC is a highly polymorphic genomic region that encodes the transplantation and immune regulatory molecules. It receives special attention for genetic investigation because of its important role in the regulation of innate and adaptive immune responses and its strong association with numerous infectious and/or autoimmune diseases. The MHC locus was first discovered in the mouse and for the past 50 years it has been studied most intensively in both mice and humans. However, in recent years the macaque species have emerged as some of the more important and advanced experimental animal models for biomedical research into MHC with important human immunodeficiency virus/simian immunodeficiency virus and transplantation studies undertaken in association with precise MHC genotyping and haplotyping methods using Sanger sequencing and next-generation sequencing. Here, in this special issue on 'Macaque Immunology' we provide a short review of the genomic similarities and differences among the human, macaque and mouse MHC class I and class II regions, with an emphasis on the association of the macaque class I region with MHC polymorphism, haplotype structure and function.
Keywords: haplotype; macaque; major histocompatibility complex; polymorphism.
© 2016 John Wiley & Sons Ltd.
Figures


Similar articles
-
The essential detail: the genetics and genomics of the primate immune response.ILAR J. 2013;54(2):181-95. doi: 10.1093/ilar/ilt043. ILAR J. 2013. PMID: 24174441 Review.
-
Discovery of novel MHC-class I alleles and haplotypes in Filipino cynomolgus macaques (Macaca fascicularis) by pyrosequencing and Sanger sequencing: Mafa-class I polymorphism.Immunogenetics. 2015 Oct;67(10):563-78. doi: 10.1007/s00251-015-0867-9. Epub 2015 Sep 8. Immunogenetics. 2015. PMID: 26349955
-
Haplessly hoping: macaque major histocompatibility complex made easy.ILAR J. 2013;54(2):196-210. doi: 10.1093/ilar/ilt036. ILAR J. 2013. PMID: 24174442 Free PMC article. Review.
-
Microsatellite typing of the rhesus macaque MHC region.Immunogenetics. 2005 May;57(3-4):198-209. doi: 10.1007/s00251-005-0787-1. Epub 2005 Apr 5. Immunogenetics. 2005. PMID: 15900491
-
A BAC-based contig map of the cynomolgus macaque (Macaca fascicularis) major histocompatibility complex genomic region.Genomics. 2007 Mar;89(3):402-12. doi: 10.1016/j.ygeno.2006.11.002. Epub 2006 Dec 14. Genomics. 2007. PMID: 17174065
Cited by
-
MHC genotyping from rhesus macaque exome sequences.Immunogenetics. 2019 Sep;71(8-9):531-544. doi: 10.1007/s00251-019-01125-w. Epub 2019 Jul 18. Immunogenetics. 2019. PMID: 31321455 Free PMC article.
-
Characterization of major histocompatibility complex-related molecule 1 sequence variants in non-human primates.Immunogenetics. 2019 Feb;71(2):109-121. doi: 10.1007/s00251-018-1091-1. Epub 2018 Oct 23. Immunogenetics. 2019. PMID: 30353260 Free PMC article.
-
Editorial: Population genomic architecture: Conserved polymorphic sequences (CPSs), not linkage disequilibrium.Front Genet. 2023 Jan 26;14:1140350. doi: 10.3389/fgene.2023.1140350. eCollection 2023. Front Genet. 2023. PMID: 36777737 Free PMC article. No abstract available.
-
Characterization of 100 extended major histocompatibility complex haplotypes in Indonesian cynomolgus macaques.Immunogenetics. 2020 May;72(4):225-239. doi: 10.1007/s00251-020-01159-5. Epub 2020 Feb 29. Immunogenetics. 2020. PMID: 32112172 Free PMC article.
-
Identification of novel polymorphisms and two distinct haplotype structures in dog leukocyte antigen class I genes: DLA-88, DLA-12 and DLA-64.Immunogenetics. 2018 Apr;70(4):237-255. doi: 10.1007/s00251-017-1031-5. Epub 2017 Sep 27. Immunogenetics. 2018. PMID: 28951951
References
-
- Zinkernagel RM, Doherty PC. The discovery of MHC restriction. Immunol Today 1997; 18:14–17. - PubMed
-
- Kulski JK, Inoko H. Major histocompatibility complex (MHC) Genes. Nat Encycl Hum Genom 2003; 3:778–85.
-
- Thorsby E. A short history of HLA. Tissue Antigens 2009; 74:101–16. - PubMed
-
- Danchin E, Vitiello V, Vienne A, Richard O, Gouret P, McDermott MF et al The major histocompatibility complex origin. Immunol Rev 2004; 198:216–32. - PubMed
-
- Joffre OP, Segura E, Savina A, Amigorena S. Cross‐presentation by dendritic cells. Nat Rev Immunol 2012; 12:557–69. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

