Comparison of various molecular methods for rapid differentiation of intestinal bifidobacteria at the species, subspecies and strain level
- PMID: 27449060
- PMCID: PMC4957357
- DOI: 10.1186/s12866-016-0779-3
Comparison of various molecular methods for rapid differentiation of intestinal bifidobacteria at the species, subspecies and strain level
Abstract
Background: Members of the genus Bifidobacterium are anaerobic Gram-positive Actinobacteria, which are natural inhabitants of human and animal gastrointestinal tract. Certain bifidobacteria are frequently used as food additives and probiotic pharmaceuticals, because of their various health-promoting properties. Due to the enormous demand on probiotic bacteria, manufacture of high-quality products containing living microorganisms requires rapid and accurate identification of specific bacteria. Additionally, isolation of new industrial bacteria from various environments may lead to multiple isolations of the same strain, therefore, it is important to apply rapid, low-cost and effective procedures differentiating bifidobacteria at the intra-species level. The identification of new isolates using microbiological and biochemical methods is difficult, but the accurate characterization of isolated strains may be achieved using a polyphasic approach that includes classical phenotypic methods and molecular procedures. However, some of these procedures are time-consuming and cumbersome, particularly when a large group of new isolates is typed, while some other approaches may have too low discriminatory power to distinguish closely related isolates obtained from similar sources.
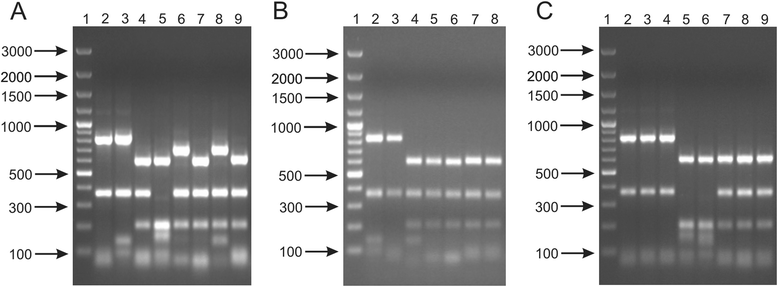
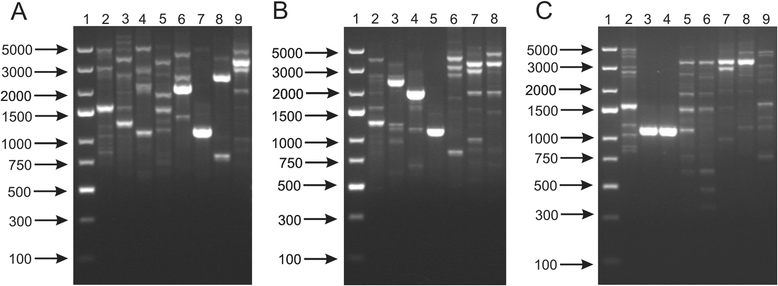
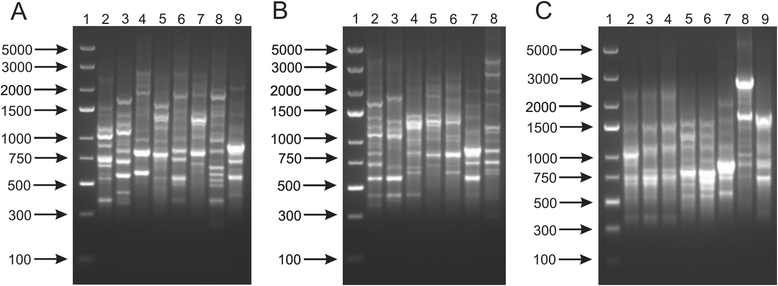
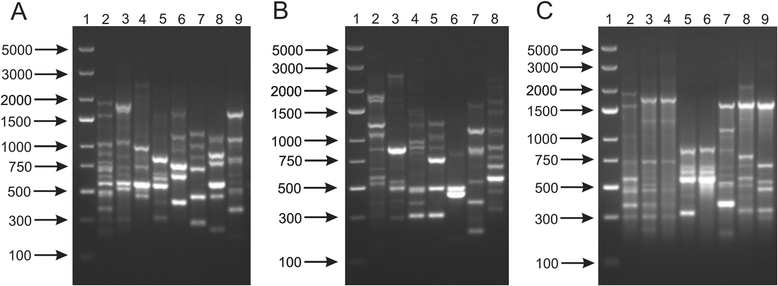
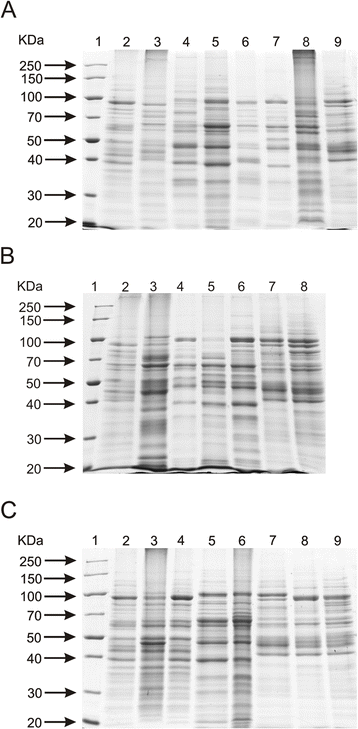
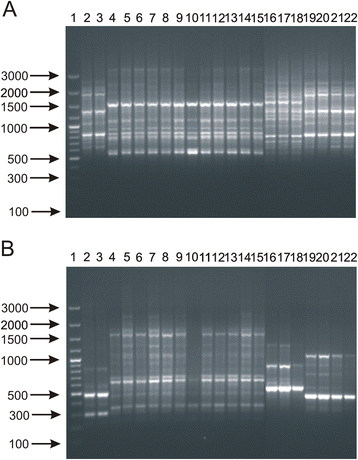
Results: This work presents the evaluation of the discriminatory power of four molecular methods (ARDRA, RAPD-PCR, rep-PCR and SDS-PAGE fingerprinting) that are extensively used for fast differentiation of bifidobacteria up to the strain level. Our experiments included 17 reference strains and showed that in comparison to ARDRA, genotypic fingerprinting procedures (RAPD and rep-PCR) seemed to be less reproducible, however, they allowed to differentiate the tested microorganisms even at the intra-species level. In general, RAPD and rep-PCR have similar discriminatory power, though, in some instances more than one oligonucleotide needs to be used in random amplified polymorphic DNA analysis. Moreover, the results also demonstrated a high discriminatory power of SDS-PAGE fingerprinting of whole-cell proteins. On the other hand, the protein profiles obtained were rather complex, and therefore, difficult to analyze.
Conclusions: Among the tested procedures, rep-PCR proved to be the most effective and reliable method allowing rapid differentiation of Bifidobacterium strains. Additionally, the use of the BOXA1R primer in the differentiation of 21 Bifidobacterium strains, newly isolated from infant feces, demonstrated slightly better discriminatory power in comparison to PCR reactions with the (GTG)5 oligonucleotide. Thus, BOX-PCR turned out to be the most appropriate and convenient molecular technique in differentiating Bifidobacterium strains at all taxonomic levels.
Keywords: ARDRA; Bifidobacterium; Differentiation; RAPD; SDS-PAGE fingerprinting; rep-PCR.
Figures
Similar articles
-
Identification of Bifidobacterium species using rep-PCR fingerprinting.Syst Appl Microbiol. 2003 Nov;26(4):557-63. doi: 10.1078/072320203770865864. Syst Appl Microbiol. 2003. PMID: 14666984
-
Efficiency of PCR-based methods in discriminating Bifidobacterium longum ssp. longum and Bifidobacterium longum ssp. infantis strains of human origin.J Microbiol Methods. 2011 Oct;87(1):10-6. doi: 10.1016/j.mimet.2011.06.014. Epub 2011 Jul 2. J Microbiol Methods. 2011. PMID: 21756944
-
[Application of the methods of molecular systematics to classification and identification of bacteria of the genus Bifidobacterium].Mikrobiologiia. 2008 May-Jun;77(3):293-302. Mikrobiologiia. 2008. PMID: 18683644 Review. Russian.
-
RAPD and rep-PCR fingerprinting for characterization of Bifidobacterium species.Folia Microbiol (Praha). 2008;53(2):99-104. doi: 10.1007/s12223-008-0014-1. Epub 2008 May 25. Folia Microbiol (Praha). 2008. PMID: 18500627
-
Molecular approaches for the detection and identification of bifidobacteria and lactobacilli in the human gastrointestinal tract.Syst Appl Microbiol. 2003 Nov;26(4):572-84. doi: 10.1078/072320203770865882. Syst Appl Microbiol. 2003. PMID: 14666986 Review.
Cited by
-
A Novel Bifidobacterium longum Subsp. longum T1 Strain from Cow's Milk: Homeostatic and Antibacterial Activity against ESBL-Producing Escherichia coli.Antibiotics (Basel). 2024 Sep 27;13(10):924. doi: 10.3390/antibiotics13100924. Antibiotics (Basel). 2024. PMID: 39452191 Free PMC article.
-
Genotyping and plant-derived glycan utilization analysis of Bifidobacterium strains from mother-infant pairs.BMC Microbiol. 2020 Sep 10;20(1):277. doi: 10.1186/s12866-020-01962-w. BMC Microbiol. 2020. PMID: 32912151 Free PMC article.
-
Human milk and mucosa-associated disaccharides impact on cultured infant fecal microbiota.Sci Rep. 2020 Jul 16;10(1):11845. doi: 10.1038/s41598-020-68718-4. Sci Rep. 2020. PMID: 32678209 Free PMC article.
-
Cultivation and Genomics Prove Long-Term Colonization of Donor's Bifidobacteria in Recurrent Clostridioides difficile Patients Treated With Fecal Microbiota Transplantation.Front Microbiol. 2020 Jul 15;11:1663. doi: 10.3389/fmicb.2020.01663. eCollection 2020. Front Microbiol. 2020. PMID: 32760391 Free PMC article.
-
Molecular typing tools for identifying and characterizing lactic acid bacteria: a review.Food Sci Biotechnol. 2020 Aug 16;29(10):1301-1318. doi: 10.1007/s10068-020-00802-x. eCollection 2020 Oct. Food Sci Biotechnol. 2020. PMID: 32995049 Free PMC article. Review.
References
-
- He T, Priebe MG, Zhong Y, Huang C, Harmsen HJM, Raangs GC, et al. Effects of yogurt and bifidobacteria supplementation on the colonic microbiota in lactose-intolerant subjects. J Appl Microbiol. 2008;104(2):595–604. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

