_targeting FLT3-ITD signaling mediates ceramide-dependent mitophagy and attenuates drug resistance in AML
- PMID: 27540013
- PMCID: PMC5064718
- DOI: 10.1182/blood-2016-04-708750
_targeting FLT3-ITD signaling mediates ceramide-dependent mitophagy and attenuates drug resistance in AML
Abstract
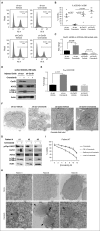
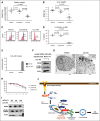
Signaling pathways regulated by mutant Fms-like tyrosine kinase 3 (FLT3)-internal tandem duplication (ITD), which mediate resistance to acute myeloid leukemia (AML) cell death, are poorly understood. Here, we reveal that pro-cell death lipid ceramide generation is suppressed by FLT3-ITD signaling. Molecular or pharmacologic inhibition of FLT3-ITD reactivated ceramide synthesis, selectively inducing mitophagy and AML cell death. Mechanistically, FLT3-ITD _targeting induced ceramide accumulation on the outer mitochondrial membrane, which then directly bound autophagy-inducing light chain 3 (LC3), involving its I35 and F52 residues, to recruit autophagosomes for execution of lethal mitophagy. Short hairpin RNA (shRNA)-mediated knockdown of LC3 prevented AML cell death in response to FLT3-ITD inhibition by crenolanib, which was restored by wild-type (WT)-LC3, but not mutants of LC3 with altered ceramide binding (I35A-LC3 or F52A-LC3). Mitochondrial ceramide accumulation and lethal mitophagy induction in response to FLT3-ITD _targeting was mediated by dynamin-related protein 1 (Drp1) activation via inhibition of protein kinase A-regulated S637 phosphorylation, resulting in mitochondrial fission. Inhibition of Drp1 prevented ceramide-dependent lethal mitophagy, and reconstitution of WT-Drp1 or phospho-null S637A-Drp1 but not its inactive phospho-mimic mutant (S637D-Drp1), restored mitochondrial fission and mitophagy in response to crenolanib in FLT3-ITD+ AML cells expressing stable shRNA against endogenous Drp1. Moreover, activating FLT3-ITD signaling in crenolanib-resistant AML cells suppressed ceramide-dependent mitophagy and prevented cell death. FLT3-ITD+ AML drug resistance is attenuated by LCL-461, a mitochondria-_targeted ceramide analog drug, in vivo, which also induced lethal mitophagy in human AML blasts with clinically relevant FLT3 mutations. Thus, these data reveal a novel mechanism which regulates AML cell death by ceramide-dependent mitophagy in response to FLT3-ITD _targeting.
© 2016 by The American Society of Hematology.
Figures

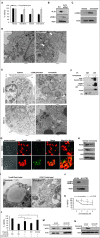
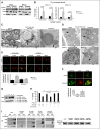
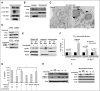
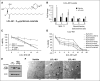
Comment in
-
Fighting fat in AML.Blood. 2016 Oct 13;128(15):1910-1911. doi: 10.1182/blood-2016-08-735704. Blood. 2016. PMID: 27737846 No abstract available.
Similar articles
-
Wu-5, a novel USP10 inhibitor, enhances crenolanib-induced FLT3-ITD-positive AML cell death via inhibiting FLT3 and AMPK pathways.Acta Pharmacol Sin. 2021 Apr;42(4):604-612. doi: 10.1038/s41401-020-0455-x. Epub 2020 Jul 21. Acta Pharmacol Sin. 2021. PMID: 32694757 Free PMC article.
-
Proteasome inhibitors induce FLT3-ITD degradation through autophagy in AML cells.Blood. 2016 Feb 18;127(7):882-92. doi: 10.1182/blood-2015-05-646497. Epub 2015 Aug 18. Blood. 2016. PMID: 26286850
-
Crenolanib is active against models of drug-resistant FLT3-ITD-positive acute myeloid leukemia.Blood. 2013 Nov 21;122(22):3607-15. doi: 10.1182/blood-2013-07-513044. Epub 2013 Sep 17. Blood. 2013. PMID: 24046014 Free PMC article.
-
FLT3 inhibitors in the treatment of acute myeloid leukemia: current status and future perspectives.Minerva Med. 2020 Oct;111(5):427-442. doi: 10.23736/S0026-4806.20.06989-X. Epub 2020 Sep 21. Minerva Med. 2020. PMID: 32955823 Review.
-
_targeting on glycosylation of mutant FLT3 in acute myeloid leukemia.Hematology. 2019 Dec;24(1):651-660. doi: 10.1080/16078454.2019.1666219. Hematology. 2019. PMID: 31533545 Review.
Cited by
-
Role of ceramides in diabetic foot ulcers (Review).Int J Mol Med. 2023 Mar;51(3):26. doi: 10.3892/ijmm.2023.5229. Epub 2023 Feb 17. Int J Mol Med. 2023. PMID: 36799149 Free PMC article. Review.
-
Sphingolipid metabolism determines the therapeutic efficacy of nanoliposomal ceramide in acute myeloid leukemia.Blood Adv. 2019 Sep 10;3(17):2598-2603. doi: 10.1182/bloodadvances.2018021295. Blood Adv. 2019. PMID: 31488436 Free PMC article.
-
Functions and Implications of Autophagy in Colon Cancer.Cells. 2019 Oct 30;8(11):1349. doi: 10.3390/cells8111349. Cells. 2019. PMID: 31671556 Free PMC article. Review.
-
Ceramide: improving Bcl-2 inhibitor therapy.Blood. 2022 Jun 30;139(26):3676-3678. doi: 10.1182/blood.2022016608. Blood. 2022. PMID: 35771560 Free PMC article. No abstract available.
-
Alterations of Mitochondria and Related Metabolic Pathways in Leukemia: A Narrative Review.Saudi J Med Med Sci. 2020 Jan-Apr;8(1):3-11. doi: 10.4103/sjmms.sjmms_112_18. Epub 2019 Dec 23. Saudi J Med Med Sci. 2020. PMID: 31929772 Free PMC article. Review.
References
-
- Kadia TM, Ravandi F, Cortes J, Kantarjian H. Toward individualized therapy in acute myeloid leukemia: a contemporary review. JAMA Oncol. 2015;1(6):820–828. - PubMed
-
- Stone RM, DeAngelo DJ, Klimek V, et al. Patients with acute myeloid leukemia and an activating mutation in FLT3 respond to a small-molecule FLT3 tyrosine kinase inhibitor, PKC412. Blood. 2005;105(1):54–60. - PubMed
-
- Kiyoi H, Ohno R, Ueda R, Saito H, Naoe T. Mechanism of constitutive activation of FLT3 with internal tandem duplication in the juxtamembrane domain. Oncogene. 2002;21(16):2555–2563. - PubMed
-
- Mizuki M, Fenski R, Halfter H, et al. Flt3 mutations from patients with acute myeloid leukemia induce transformation of 32D cells mediated by the Ras and STAT5 pathways. Blood. 2000;96(12):3907–3914. - PubMed
-
- Choudhary C, Schwäble J, Brandts C, et al. AML-associated Flt3 kinase domain mutations show signal transduction differences compared with Flt3 ITD mutations. Blood. 2005;106(1):265–273. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

