Structural Insights into Histone Crotonyl-Lysine Recognition by the AF9 YEATS Domain
- PMID: 27545619
- PMCID: PMC5014688
- DOI: 10.1016/j.str.2016.05.023
Structural Insights into Histone Crotonyl-Lysine Recognition by the AF9 YEATS Domain
Abstract
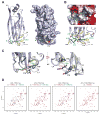
Histone lysine acylations play an important role in the regulation of gene transcription in chromatin. Unlike histone acetyl-lysine, molecular recognition of a recently identified crotonyl-lysine mark is much less understood. Here, we report that the YEATS domain of AF9 preferentially binds crotonyl-lysine over acetyl-lysine in histone H3. Nuclear magnetic resonance structural analysis reveals that crotonyl-lysine of histone H3 lysine 18 is engulfed deep in an aromatic cage of the YEATS domain where the carbonyl oxygen of crotonyl-lysine forms a hydrogen bond with the backbone amide of protein residue Tyr78. The crotonyl-lysine, through its unique electron-rich double-bond side chain, engages π-π aromatic stacking and extended hydrophobic/aromatic interactions with the YEATS domain compared with acetyl-lysine. Our mutational analysis confirmed key protein residues Phe59 and Tyr78 for crotonyl-lysine recognition. Importantly, our findings present a new structural mechanism of protein-protein interactions mediated by histone lysine crotonylation, and show how the cells interpret acyl-lysine marks in different biological contexts.
Copyright © 2016 Elsevier Ltd. All rights reserved.
Conflict of interest statement
STATEMENT The authors declare no competing financial interests.
Figures

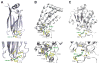
Similar articles
-
Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain.Mol Cell. 2016 Apr 21;62(2):181-193. doi: 10.1016/j.molcel.2016.03.028. Mol Cell. 2016. PMID: 27105114 Free PMC article.
-
Structural insights into the π-π-π stacking mechanism and DNA-binding activity of the YEATS domain.Nat Commun. 2018 Nov 1;9(1):4574. doi: 10.1038/s41467-018-07072-6. Nat Commun. 2018. PMID: 30385749 Free PMC article.
-
Structural and mechanistic insights into regulation of HBO1 histone acetyltransferase activity by BRPF2.Nucleic Acids Res. 2017 Jun 2;45(10):5707-5719. doi: 10.1093/nar/gkx142. Nucleic Acids Res. 2017. PMID: 28334966 Free PMC article.
-
YEATS Domain-A Histone Acylation Reader in Health and Disease.J Mol Biol. 2017 Jun 30;429(13):1994-2002. doi: 10.1016/j.jmb.2017.03.010. Epub 2017 Mar 11. J Mol Biol. 2017. PMID: 28300602 Review.
-
Structural and sequence motifs of protein (histone) methylation enzymes.Annu Rev Biophys Biomol Struct. 2005;34:267-94. doi: 10.1146/annurev.biophys.34.040204.144452. Annu Rev Biophys Biomol Struct. 2005. PMID: 15869391 Free PMC article. Review.
Cited by
-
Histone Readers and Their Roles in Cancer.Cancer Treat Res. 2023;190:245-272. doi: 10.1007/978-3-031-45654-1_8. Cancer Treat Res. 2023. PMID: 38113004 Free PMC article.
-
Fragment-Based Discovery of AF9 YEATS Domain Inhibitors.Int J Mol Sci. 2022 Mar 31;23(7):3893. doi: 10.3390/ijms23073893. Int J Mol Sci. 2022. PMID: 35409252 Free PMC article.
-
Chromatin dynamics and histone modifications in intestinal microbiota-host crosstalk.Mol Metab. 2020 Aug;38:100925. doi: 10.1016/j.molmet.2019.12.005. Epub 2019 Dec 27. Mol Metab. 2020. PMID: 31992511 Free PMC article. Review.
-
Metabolic regulation of gene expression through histone acylations.Nat Rev Mol Cell Biol. 2017 Feb;18(2):90-101. doi: 10.1038/nrm.2016.140. Epub 2016 Dec 7. Nat Rev Mol Cell Biol. 2017. PMID: 27924077 Free PMC article. Review.
-
Protein lysine crotonylation: past, present, perspective.Cell Death Dis. 2021 Jul 14;12(7):703. doi: 10.1038/s41419-021-03987-z. Cell Death Dis. 2021. PMID: 34262024 Free PMC article. Review.
References
-
- Brünger ATAP, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. - PubMed
-
- Clore GM, Gronenborn AM. Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods in enzymology. 1994;239:349–363. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

