GOLM1 Modulates EGFR/RTK Cell-Surface Recycling to Drive Hepatocellular Carcinoma Metastasis
- PMID: 27569582
- PMCID: PMC5021625
- DOI: 10.1016/j.ccell.2016.07.017
GOLM1 Modulates EGFR/RTK Cell-Surface Recycling to Drive Hepatocellular Carcinoma Metastasis
Abstract
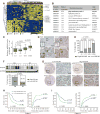
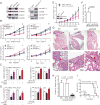
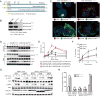
The mechanism of cancer metastasis remains poorly understood. Using gene profiling of hepatocellular carcinoma (HCC) tissues, we have identified GOLM1 as a leading gene relating to HCC metastasis. GOLM1 expression is correlated with early recurrence, metastasis, and poor survival of HCC patients. Both gain- and loss-of-function studies determine that GOLM1 acts as a key oncogene by promoting HCC growth and metastasis. It selectively interacts with epidermal growth factor receptor (EGFR) and serves as a specific cargo adaptor to assist EGFR/RTK anchoring on the trans-Golgi network (TGN) and recycling back to the plasma membrane, leading to prolonged activation of the downstream kinases. These findings reveal the functional role of GOLM1, a Golgi-related protein, in EGFR/RTK recycling and metastatic progression of HCC.
Copyright © 2016 Elsevier Inc. All rights reserved.
Conflict of interest statement
The author(s) indicated no potential conflicts of interest.
Figures




Comment in
-
Powering Tumor Metastasis with Recycled Fuel.Cancer Cell. 2016 Sep 12;30(3):374-375. doi: 10.1016/j.ccell.2016.08.014. Cancer Cell. 2016. PMID: 27622330
Similar articles
-
GOLM1 dictates acquired Lenvatinib resistance by a GOLM1-CSN5 positive feedback loop upon EGFR signaling activation in hepatocellular carcinoma.Oncogene. 2024 Oct;43(42):3108-3120. doi: 10.1038/s41388-024-03153-7. Epub 2024 Sep 9. Oncogene. 2024. PMID: 39251847
-
GOLM1-regulated EGFR/RTK recycling is a novel _target for combating HCC metastasis.Sci China Life Sci. 2017 Jan;60(1):98-101. doi: 10.1007/s11427-016-0311-x. Epub 2016 Nov 17. Sci China Life Sci. 2017. PMID: 27858335 No abstract available.
-
Liver fibrosis promotes immune escape in hepatocellular carcinoma via GOLM1-mediated PD-L1 upregulation.Cancer Lett. 2021 Aug 10;513:14-25. doi: 10.1016/j.canlet.2021.05.007. Epub 2021 May 14. Cancer Lett. 2021. PMID: 33992711
-
Recent advances of GOLM1 in hepatocellular carcinoma.Hepat Oncol. 2020 Jun 29;7(2):HEP22. doi: 10.2217/hep-2020-0006. Hepat Oncol. 2020. PMID: 32647567 Free PMC article. Review.
-
Unveiling the impact of GOLM1/GP73 on cytokine production in cancer and infectious disease.Immunol Cell Biol. 2023 Sep;101(8):727-734. doi: 10.1111/imcb.12664. Epub 2023 Jun 18. Immunol Cell Biol. 2023. PMID: 37332154 Review.
Cited by
-
Updating the Clinical Application of Blood Biomarkers and Their Algorithms in the Diagnosis and Surveillance of Hepatocellular Carcinoma: A Critical Review.Int J Mol Sci. 2023 Feb 21;24(5):4286. doi: 10.3390/ijms24054286. Int J Mol Sci. 2023. PMID: 36901717 Free PMC article. Review.
-
Knockdown of Golgi phosphoprotein 73 blocks the trafficking of matrix metalloproteinase-2 in hepatocellular carcinoma cells and inhibits cell invasion.J Cell Mol Med. 2019 Apr;23(4):2399-2409. doi: 10.1111/jcmm.14055. Epub 2019 Jan 24. J Cell Mol Med. 2019. PMID: 30677226 Free PMC article.
-
A low amino acid environment promotes cell macropinocytosis through the YY1-FGD6 axis in Ras-mutant pancreatic ductal adenocarcinoma.Oncogene. 2022 Feb;41(8):1203-1215. doi: 10.1038/s41388-021-02159-9. Epub 2022 Jan 27. Oncogene. 2022. PMID: 35082383
-
A Systems Approach to Interrogate Gene Expression Patterns in African American Men Presenting with Clinically Localized Prostate Cancer.Cancers (Basel). 2021 Oct 14;13(20):5143. doi: 10.3390/cancers13205143. Cancers (Basel). 2021. PMID: 34680291 Free PMC article.
-
GCC2 promotes non-small cell lung cancer progression by maintaining Golgi apparatus integrity and stimulating EGFR signaling pathways.Sci Rep. 2024 Nov 22;14(1):28926. doi: 10.1038/s41598-024-75316-1. Sci Rep. 2024. PMID: 39572606 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

