Early Growth Response 1 (Egr-1) Is a Transcriptional Activator of β-Secretase 1 (BACE-1) in the Brain
- PMID: 27576688
- PMCID: PMC5064006
- DOI: 10.1074/jbc.M116.738849
Early Growth Response 1 (Egr-1) Is a Transcriptional Activator of β-Secretase 1 (BACE-1) in the Brain
Abstract
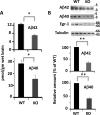
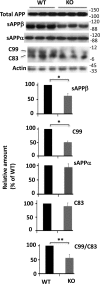
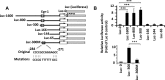
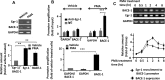
Accumulation of amyloid-β peptide (Aβ) in the brain is regarded as central to Alzheimer's disease (AD) pathogenesis. Aβ is generated by a sequential cleavage of amyloid precursor protein (APP) by β-secretase 1 (BACE-1) followed by γ-secretase. BACE-1 cleavage of APP is the committed step in Aβ synthesis. Understanding the mechanism by which BACE-1 is activated leading to Aβ synthesis in the brain can provide better understanding of AD pathology and help to develop novel therapies. In this study, we found that the levels of Aβ and BACE-1 are significantly reduced in the brains of mice lacking transcription factor early growth response 1 (Egr-1) when compared with the WT. We demonstrate that in COS-7 cells, Egr-1 binds to the BACE-1 promoter and activates BACE-1 transcription. In rat hippocampal primary neurons, overexpression of Egr-1 induces BACE-1 expression, activates BACE-1, promotes amyloidogenic APP processing, and enhances Aβ synthesis. In mouse hippocampal primary neurons, knockdown of BACE-1 almost completely blocks Egr-1-induced amyloidogenic APP processing and Aβ synthesis. Our data indicate that Egr-1 promotes Aβ synthesis via transcriptional activation of BACE-1 and suggest that Egr-1 plays role in activation of BACE-1 and acceleration of Aβ synthesis in AD brain. Egr-1 is a potential therapeutic _target for AD.
Keywords: amyloid-β (Aβ); early growth response protein 1 (EGR1); gene regulation; neuroscience; promoter; transcription factor.
© 2016 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures



Similar articles
-
Inhibition of Early Growth Response 1 in the Hippocampus Alleviates Neuropathology and Improves Cognition in an Alzheimer Model with Plaques and Tangles.Am J Pathol. 2017 Aug;187(8):1828-1847. doi: 10.1016/j.ajpath.2017.04.018. Epub 2017 Jun 20. Am J Pathol. 2017. PMID: 28641077
-
Beta-secretase cleavage at amino acid residue 34 in the amyloid beta peptide is dependent upon gamma-secretase activity.J Biol Chem. 2003 Jun 6;278(23):21286-94. doi: 10.1074/jbc.M209859200. Epub 2003 Mar 27. J Biol Chem. 2003. PMID: 12665519
-
BACE knockout mice are healthy despite lacking the primary beta-secretase activity in brain: implications for Alzheimer's disease therapeutics.Hum Mol Genet. 2001 Jun 1;10(12):1317-24. doi: 10.1093/hmg/10.12.1317. Hum Mol Genet. 2001. PMID: 11406613
-
Alzheimer's disease.Subcell Biochem. 2012;65:329-52. doi: 10.1007/978-94-007-5416-4_14. Subcell Biochem. 2012. PMID: 23225010 Review.
-
The beta-secretase, BACE: a prime drug _target for Alzheimer's disease.J Mol Neurosci. 2001 Oct;17(2):157-70. doi: 10.1385/JMN:17:2:157. J Mol Neurosci. 2001. PMID: 11816789 Review.
Cited by
-
Clusterin Is Required for β-Amyloid Toxicity in Human iPSC-Derived Neurons.Front Neurosci. 2018 Jul 25;12:504. doi: 10.3389/fnins.2018.00504. eCollection 2018. Front Neurosci. 2018. PMID: 30090055 Free PMC article.
-
Sex differences in hippocampal β-amyloid accumulation in the triple-transgenic mouse model of Alzheimer's disease and the potential role of local estrogens.Front Neurosci. 2023 Mar 8;17:1117584. doi: 10.3389/fnins.2023.1117584. eCollection 2023. Front Neurosci. 2023. PMID: 36968493 Free PMC article.
-
Comparative Transcriptome Analysis in Monocyte-Derived Macrophages of Asymptomatic GBA Mutation Carriers and Patients with GBA-Associated Parkinson's Disease.Genes (Basel). 2021 Sep 29;12(10):1545. doi: 10.3390/genes12101545. Genes (Basel). 2021. PMID: 34680941 Free PMC article.
-
A Novel Egr-1-Agrin Pathway and Potential Implications for Regulation of Synaptic Physiology and Homeostasis at the Neuromuscular Junction.Front Aging Neurosci. 2017 Aug 3;9:258. doi: 10.3389/fnagi.2017.00258. eCollection 2017. Front Aging Neurosci. 2017. PMID: 28824419 Free PMC article.
-
Krüppel-like factor 5 accelerates the pathogenesis of Alzheimer's disease via BACE1-mediated APP processing.Alzheimers Res Ther. 2022 Jul 26;14(1):103. doi: 10.1186/s13195-022-01050-3. Alzheimers Res Ther. 2022. PMID: 35883144 Free PMC article.
References
-
- Yang L. B., Lindholm K., Yan R., Citron M., Xia W., Yang X. L., Beach T., Sue L., Wong P., Price D., Li R., and Shen Y. (2003) Elevated β-secretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat. Med. 9, 3–4 - PubMed
-
- Fukumoto H., Cheung B. S., Hyman B. T., and Irizarry M. C. (2002) β-Secretase protein and activity are increased in the neocortex in Alzheimer disease. Arch. Neurol. 59, 1381–1389 - PubMed
-
- Holsinger R. M., McLean C. A., Beyreuther K., Masters C. L., and Evin G. (2002) Increased expression of the amyloid precursor β-secretase in Alzheimer's disease. Ann. Neurol. 51, 783–786 - PubMed
-
- Tyler S. J., Dawbarn D., Wilcock G. K., and Allen S. J. (2002) α- and β-secretase: profound changes in Alzheimer's disease. Biochem. Biophys. Res. Commun. 299, 373–376 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

