Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition
- PMID: 27580029
- PMCID: PMC5136292
- DOI: 10.1038/nature19353
Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition
Erratum in
-
Corrigendum: Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition.Nature. 2016 Dec 1;540(7631):150. doi: 10.1038/nature20144. Epub 2016 Oct 19. Nature. 2016. PMID: 27760119 No abstract available.
Abstract
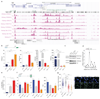
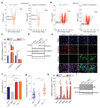
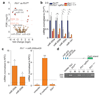
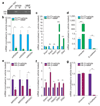
Mutations of the tricarboxylic acid cycle enzyme fumarate hydratase cause hereditary leiomyomatosis and renal cell cancer. Fumarate hydratase-deficient renal cancers are highly aggressive and metastasize even when small, leading to a very poor clinical outcome. Fumarate, a small molecule metabolite that accumulates in fumarate hydratase-deficient cells, plays a key role in cell transformation, making it a bona fide oncometabolite. Fumarate has been shown to inhibit α-ketoglutarate-dependent dioxygenases that are involved in DNA and histone demethylation. However, the link between fumarate accumulation, epigenetic changes, and tumorigenesis is unclear. Here we show that loss of fumarate hydratase and the subsequent accumulation of fumarate in mouse and human cells elicits an epithelial-to-mesenchymal-transition (EMT), a phenotypic switch associated with cancer initiation, invasion, and metastasis. We demonstrate that fumarate inhibits Tet-mediated demethylation of a regulatory region of the antimetastatic miRNA cluster mir-200ba429, leading to the expression of EMT-related transcription factors and enhanced migratory properties. These epigenetic and phenotypic changes are recapitulated by the incubation of fumarate hydratase-proficient cells with cell-permeable fumarate. Loss of fumarate hydratase is associated with suppression of miR-200 and the EMT signature in renal cancer and is associated with poor clinical outcome. These results imply that loss of fumarate hydratase and fumarate accumulation contribute to the aggressive features of fumarate hydratase-deficient tumours.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
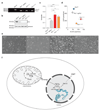
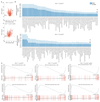

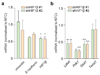

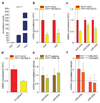




Comment in
-
Oncometabolite Accumulation and Epithelial-to-Mesenchymal Transition: The Turn of Fumarate.Cell Metab. 2016 Oct 11;24(4):529-530. doi: 10.1016/j.cmet.2016.09.020. Cell Metab. 2016. PMID: 27732835
Similar articles
-
Fumarates and Cancer.Trends Mol Med. 2017 Jan;23(1):3-5. doi: 10.1016/j.molmed.2016.12.001. Epub 2016 Dec 13. Trends Mol Med. 2017. PMID: 27986420
-
Oncometabolite Accumulation and Epithelial-to-Mesenchymal Transition: The Turn of Fumarate.Cell Metab. 2016 Oct 11;24(4):529-530. doi: 10.1016/j.cmet.2016.09.020. Cell Metab. 2016. PMID: 27732835
-
Fumarate Hydratase Loss Causes Combined Respiratory Chain Defects.Cell Rep. 2017 Oct 24;21(4):1036-1047. doi: 10.1016/j.celrep.2017.09.092. Cell Rep. 2017. PMID: 29069586 Free PMC article.
-
Functional implications of fumarate-induced cysteine succination.Trends Biochem Sci. 2024 Sep;49(9):775-790. doi: 10.1016/j.tibs.2024.05.003. Epub 2024 Jun 13. Trends Biochem Sci. 2024. PMID: 38876954 Review.
-
Fumarate hydratase inactivation in renal tumors: HIF1α, NRF2, and "cryptic _targets" of transcription factors.Chin J Cancer. 2012 Sep;31(9):413-20. doi: 10.5732/cjc.012.10102. Epub 2012 Jul 2. Chin J Cancer. 2012. PMID: 22776233 Free PMC article. Review.
Cited by
-
Tumor Cell-Intrinsic Immunometabolism and Precision Nutrition in Cancer Immunotherapy.Cancers (Basel). 2020 Jul 2;12(7):1757. doi: 10.3390/cancers12071757. Cancers (Basel). 2020. PMID: 32630618 Free PMC article.
-
A break in mitochondrial endosymbiosis as a basis for inflammatory diseases.Nature. 2024 Feb;626(7998):271-279. doi: 10.1038/s41586-023-06866-z. Epub 2024 Feb 7. Nature. 2024. PMID: 38326590 Review.
-
Nitrogen Trapping as a Therapeutic Strategy in Tumors with Mitochondrial Dysfunction.Cancer Res. 2020 Sep 1;80(17):3492-3506. doi: 10.1158/0008-5472.CAN-20-0246. Epub 2020 Jul 10. Cancer Res. 2020. PMID: 32651261 Free PMC article.
-
Mitochondrial metabolism and cancer metastasis.Ann Transl Med. 2020 Jul;8(14):904. doi: 10.21037/atm.2020.03.42. Ann Transl Med. 2020. PMID: 32793748 Free PMC article. Review.
-
Malic enzyme 2 connects the Krebs cycle intermediate fumarate to mitochondrial biogenesis.Cell Metab. 2021 May 4;33(5):1027-1041.e8. doi: 10.1016/j.cmet.2021.03.003. Epub 2021 Mar 25. Cell Metab. 2021. PMID: 33770508 Free PMC article.
References
-
- Tomlinson IP, Alam NA, Rowan AJ, Barclay E, Jaeger EE, Kelsell D, Leigh I, Gorman P, Lamlum H, Rahman S, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet. 2002;30:406–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

