Autophagy requires poly(adp-ribosyl)ation-dependent AMPK nuclear export
- PMID: 27689873
- PMCID: PMC5136490
- DOI: 10.1038/cdd.2016.80
Autophagy requires poly(adp-ribosyl)ation-dependent AMPK nuclear export
Abstract
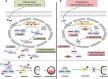
AMPK is a central energy sensor linking extracellular milieu fluctuations with the autophagic machinery. In the current study we uncover that Poly(ADP-ribosyl)ation (PARylation), a post-translational modification (PTM) of proteins, accounts for the spatial and temporal regulation of autophagy by modulating AMPK subcellular localisation and activation. More particularly, we show that the minority AMPK pool needs to be exported to the cytosol in a PARylation-dependent manner for optimal induction of autophagy, including ULK1 phosphorylation and mTORC1 inactivation. PARP-1 forms a molecular complex with AMPK in the nucleus in non-starved cells. In response to nutrient deprivation, PARP-1 catalysed PARylation, induced the dissociation of the PARP-1/AMPK complex and the export of free PARylated nuclear AMPK to the cytoplasm to activate autophagy. PARP inhibition, its silencing or the expression of PARylation-deficient AMPK mutants prevented not only the AMPK nuclear-cytosolic export but also affected the activation of the cytosolic AMPK pool and autophagosome formation. These results demonstrate that PARylation of AMPK is a key early signal to efficiently convey extracellular nutrient perturbations with downstream events needed for the cell to optimize autophagic commitment before autophagosome formation.
Figures






Similar articles
-
Regulation of NFAT by poly(ADP-ribose) polymerase activity in T cells.Mol Immunol. 2008 Apr;45(7):1863-71. doi: 10.1016/j.molimm.2007.10.044. Mol Immunol. 2008. PMID: 18078995
-
Fine-tuning of AMPK-ULK1-mTORC1 regulatory triangle is crucial for autophagy oscillation.Sci Rep. 2020 Oct 20;10(1):17803. doi: 10.1038/s41598-020-75030-8. Sci Rep. 2020. PMID: 33082544 Free PMC article.
-
Poly-ADP ribosylation in DNA damage response and cancer therapy.Mutat Res Rev Mutat Res. 2019 Apr-Jun;780:82-91. doi: 10.1016/j.mrrev.2017.09.004. Epub 2017 Sep 20. Mutat Res Rev Mutat Res. 2019. PMID: 31395352 Free PMC article. Review.
-
AMPK Inhibits ULK1-Dependent Autophagosome Formation and Lysosomal Acidification via Distinct Mechanisms.Mol Cell Biol. 2018 Apr 30;38(10):e00023-18. doi: 10.1128/MCB.00023-18. Print 2018 May 15. Mol Cell Biol. 2018. PMID: 29507183 Free PMC article.
-
Poly(ADP-ribose) signaling in cell death.Mol Aspects Med. 2013 Dec;34(6):1153-67. doi: 10.1016/j.mam.2013.01.007. Epub 2013 Feb 15. Mol Aspects Med. 2013. PMID: 23416893 Review.
Cited by
-
Understanding the Role of Autophagy in Cancer Formation and Progression Is a Real Opportunity to Treat and Cure Human Cancers.Cancers (Basel). 2021 Nov 10;13(22):5622. doi: 10.3390/cancers13225622. Cancers (Basel). 2021. PMID: 34830777 Free PMC article. Review.
-
Thrap3 promotes nonalcoholic fatty liver disease by suppressing AMPK-mediated autophagy.Exp Mol Med. 2023 Aug;55(8):1720-1733. doi: 10.1038/s12276-023-01047-4. Epub 2023 Aug 1. Exp Mol Med. 2023. PMID: 37524868 Free PMC article.
-
DIRAS3-Derived Peptide Inhibits Autophagy in Ovarian Cancer Cells by Binding to Beclin1.Cancers (Basel). 2019 Apr 18;11(4):557. doi: 10.3390/cancers11040557. Cancers (Basel). 2019. PMID: 31003488 Free PMC article.
-
Metabolic Reprogramming in Glioma.Front Cell Dev Biol. 2017 Apr 26;5:43. doi: 10.3389/fcell.2017.00043. eCollection 2017. Front Cell Dev Biol. 2017. PMID: 28491867 Free PMC article. Review.
-
Human Nmnat1 Promotes Autophagic Clearance of Amyloid Plaques in a Drosophila Model of Alzheimer's Disease.Front Aging Neurosci. 2022 Mar 24;14:852972. doi: 10.3389/fnagi.2022.852972. eCollection 2022. Front Aging Neurosci. 2022. PMID: 35401143 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

