Gene expression profiling of epigenetic chromatin modification enzymes and histone marks by cigarette smoke: implications for COPD and lung cancer
- PMID: 27793800
- PMCID: PMC5206398
- DOI: 10.1152/ajplung.00253.2016
Gene expression profiling of epigenetic chromatin modification enzymes and histone marks by cigarette smoke: implications for COPD and lung cancer
Abstract
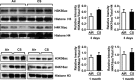
Chromatin-modifying enzymes mediate DNA methylation and histone modifications on recruitment to specific _target gene loci in response to various stimuli. The key enzymes that regulate chromatin accessibility for maintenance of modifications in DNA and histones, and for modulation of gene expression patterns in response to cigarette smoke (CS), are not known. We hypothesize that CS exposure alters the gene expression patterns of chromatin-modifying enzymes, which then affects multiple downstream pathways involved in the response to CS. We have, therefore, analyzed chromatin-modifying enzyme profiles and validated by quantitative real-time PCR (qPCR). We also performed immunoblot analysis of _targeted histone marks in C57BL/6J mice exposed to acute and subchronic CS, and of lungs from nonsmokers, smokers, and patients with chronic obstructive pulmonary disease (COPD). We found a significant increase in expression of several chromatin modification enzymes, including DNA methyltransferases, histone acetyltransferases, histone methyltransferases, and SET domain proteins, histone kinases, and ubiquitinases. Our qPCR validation data revealed a significant downregulation of Dnmt1, Dnmt3a, Dnmt3b, Hdac2, Hdac4, Hat1, Prmt1, and Aurkb We identified _targeted chromatin histone marks (H3K56ac and H4K12ac), which are induced by CS. Thus CS-induced genotoxic stress differentially affects the expression of epigenetic modulators that regulate transcription of _target genes via DNA methylation and site-specific histone modifications. This may have implications in devising epigenetic-based therapies for COPD and lung cancer.
Keywords: COPD; chromatin modifications; cigarette smoke; epigenetics; epithelial cells; lungs.
Copyright © 2016 the American Physiological Society.
Figures


Similar articles
-
Cigarette smoke induces distinct histone modifications in lung cells: implications for the pathogenesis of COPD and lung cancer.J Proteome Res. 2014 Feb 7;13(2):982-96. doi: 10.1021/pr400998n. Epub 2013 Dec 13. J Proteome Res. 2014. PMID: 24283195 Free PMC article.
-
Current perspectives on role of chromatin modifications and deacetylases in lung inflammation in COPD.COPD. 2009 Aug;6(4):291-7. doi: 10.1080/15412550903049132. COPD. 2009. PMID: 19811389 Free PMC article. Review.
-
Oxidative stress and chromatin remodeling in chronic obstructive pulmonary disease and smoking-related diseases.Antioxid Redox Signal. 2013 May 20;18(15):1956-71. doi: 10.1089/ars.2012.4863. Epub 2012 Nov 6. Antioxid Redox Signal. 2013. PMID: 22978694 Free PMC article. Review.
-
The "Epigenetic Code Replication Machinery", ECREM: a promising drugable _target of the epigenetic cell memory.Curr Med Chem. 2007;14(25):2629-41. doi: 10.2174/092986707782023244. Curr Med Chem. 2007. PMID: 17979715 Review.
-
Lung cancer and its association with chronic obstructive pulmonary disease: update on nexus of epigenetics.Curr Opin Pulm Med. 2011 Jul;17(4):279-85. doi: 10.1097/MCP.0b013e3283477533. Curr Opin Pulm Med. 2011. PMID: 21537190 Free PMC article. Review.
Cited by
-
Current views in chronic obstructive pulmonary disease pathogenesis and management.Saudi Pharm J. 2021 Dec;29(12):1361-1373. doi: 10.1016/j.jsps.2021.10.008. Epub 2021 Oct 29. Saudi Pharm J. 2021. PMID: 35002373 Free PMC article. Review.
-
Epigenetic impacts of maternal tobacco and e-vapour exposure on the offspring lung.Clin Epigenetics. 2019 Feb 19;11(1):32. doi: 10.1186/s13148-019-0631-3. Clin Epigenetics. 2019. PMID: 30782202 Free PMC article. Review.
-
The role of cigarette smoke-induced pulmonary vascular endothelial cell apoptosis in COPD.Respir Res. 2021 Feb 5;22(1):39. doi: 10.1186/s12931-021-01630-1. Respir Res. 2021. PMID: 33546691 Free PMC article. Review.
-
Prenatal smoke exposure induces persistent Cyp2a5 methylation and increases nicotine metabolism in the liver of neonatal and adult male offspring.Epigenetics. 2020 Dec;15(12):1370-1385. doi: 10.1080/15592294.2020.1782655. Epub 2020 Jun 23. Epigenetics. 2020. PMID: 32573327 Free PMC article.
-
In utero exposure to electronic-cigarette aerosols decreases lung fibrillar collagen content, increases Newtonian resistance and induces sex-specific molecular signatures in neonatal mice.Toxicol Res. 2021 Sep 6;38(2):205-224. doi: 10.1007/s43188-021-00103-3. eCollection 2022 Apr. Toxicol Res. 2021. PMID: 35415078 Free PMC article.
References
-
- Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev 12: 142–148, 2002. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

