Identification of Reference Genes for Quantitative Real-Time PCR in Date Palm (Phoenix dactylifera L.) Subjected to Drought and Salinity
- PMID: 27824922
- PMCID: PMC5100987
- DOI: 10.1371/journal.pone.0166216
Identification of Reference Genes for Quantitative Real-Time PCR in Date Palm (Phoenix dactylifera L.) Subjected to Drought and Salinity
Abstract
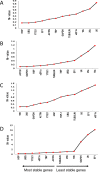
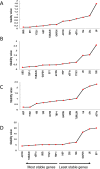
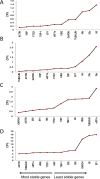
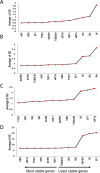
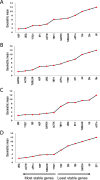
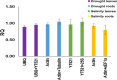
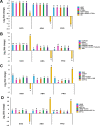
Date palm is an important crop plant in the arid and semi-arid regions supporting human population in the Middle East and North Africa. These areas have been largely affected by drought and salinity due to insufficient rainfall and improper irrigation practices. Date palm is a relatively salt- and drought-tolerant plant and more recently efforts have been directed to identifying genes and pathways that confer stress tolerance in this species. Quantitative real-time PCR (qPCR) is a promising technique for the analysis of stress-induced differential gene expression, which involves the use of stable reference genes for normalizing gene expression. In an attempt to find the best reference genes for date palm's drought and salinity research, we evaluated the stability of 12 most commonly used reference genes using the geNorm, NormFinder, BestKeeper statistical algorithms and the comparative ΔCT method. The comprehensive results revealed that HEAT SHOCK PROTEIN (HSP), UBIQUITIN (UBQ) and YTH domain-containing family protein (YT521) were stable in drought-stressed leaves whereas GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (GAPDH), ACTIN and TUBULIN were stable in drought-stressed roots. On the other hand, SMALL SUBUNIT RIBOSOMAL RNA (25S), YT521 and 18S ribosomal RNA (18S); and UBQ, ACTIN and ELONGATION FACTOR 1-ALPHA (eEF1a) were stable in leaves and roots, respectively, under salt stress. The stability of these reference genes was verified by using the abiotic stress-responsive CYTOSOLIC Cu/Zn SUPEROXIDE DISMUTASE (Cyt-Cu/Zn SOD), an ABA RECEPTOR, and a PROLINE TRANSPORTER 2 (PRO) genes. A combination of top 2 or 3 stable reference genes were found to be suitable for normalization of the _target gene expression and will facilitate gene expression analysis studies aimed at identifying functional genes associated with drought and salinity tolerance in date palm.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Genome-wide expression profiling in leaves and roots of date palm (Phoenix dactylifera L.) exposed to salinity.BMC Genomics. 2017 Mar 22;18(1):246. doi: 10.1186/s12864-017-3633-6. BMC Genomics. 2017. PMID: 28330456 Free PMC article.
-
Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Mulberry (Morus alba L.) under different abiotic stresses.Mol Biol Rep. 2019 Apr;46(2):1809-1817. doi: 10.1007/s11033-019-04631-y. Epub 2019 Jan 29. Mol Biol Rep. 2019. PMID: 30694457
-
Identification of Candidate Genes Involved in the Salt Tolerance of Date Palm (Phoenix dactylifera L.) Based on a Yeast Functional Bioassay.DNA Cell Biol. 2018 Jun;37(6):524-534. doi: 10.1089/dna.2018.4159. Epub 2018 Mar 29. DNA Cell Biol. 2018. PMID: 29596001
-
Prospects for the Study and Improvement of Abiotic Stress Tolerance in Date Palms in the Post-genomics Era.Front Plant Sci. 2020 Mar 17;11:293. doi: 10.3389/fpls.2020.00293. eCollection 2020. Front Plant Sci. 2020. PMID: 32256513 Free PMC article. Review.
-
Plant reference genes for development and stress response studies.J Biosci. 2018 Mar;43(1):173-187. J Biosci. 2018. PMID: 29485125 Review.
Cited by
-
Genome-wide expression profiling in leaves and roots of date palm (Phoenix dactylifera L.) exposed to salinity.BMC Genomics. 2017 Mar 22;18(1):246. doi: 10.1186/s12864-017-3633-6. BMC Genomics. 2017. PMID: 28330456 Free PMC article.
-
Overexpression of a Metallothionein 2A Gene from Date Palm Confers Abiotic Stress Tolerance to Yeast and Arabidopsis thaliana.Int J Mol Sci. 2019 Jun 12;20(12):2871. doi: 10.3390/ijms20122871. Int J Mol Sci. 2019. PMID: 31212812 Free PMC article.
-
Identification and Validation of Reference Genes for RT-qPCR Analysis in Switchgrass under Heavy Metal Stresses.Genes (Basel). 2020 May 3;11(5):502. doi: 10.3390/genes11050502. Genes (Basel). 2020. PMID: 32375288 Free PMC article.
-
Identification of Reliable Reference Genes under Different Stresses and in Different Tissues of Toxicodendron succedaneum.Genes (Basel). 2022 Dec 17;13(12):2396. doi: 10.3390/genes13122396. Genes (Basel). 2022. PMID: 36553662 Free PMC article.
-
Identification and Validation of Reference Genes for Gene Expression Analysis in Schima superba.Genes (Basel). 2021 May 13;12(5):732. doi: 10.3390/genes12050732. Genes (Basel). 2021. PMID: 34068362 Free PMC article.
References
-
- Chao CT, Krueger RR. The date palm (Phoenix dactylifera L.): Overview of biology, uses, and cultivation. HortScience. 2007; 42: 1077–1082.
-
- Al-Yahyai R, Khan MM. Date Palm Status and Perspective in Oman In: Al-Khayri JM, Jain SM, Johnson DV, editors. Date palm genetic resources and utilization. Netherlands: Springer Science and Business Media; 2015. pp. 207–240. 10.1007/978-94-017-9707-8_6 - DOI
-
- El-Juhany LI. Degradation of date palm trees and date production in Arab countries: causes and potential rehabilitation. Aust J Basic Appl Sci. 2010; 4: 3998–4010.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

