The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins
- PMID: 27827996
- PMCID: PMC5133849
- DOI: 10.3390/ijms17111849
The Bromodomain and Extra-Terminal Domain (BET) Family: Functional Anatomy of BET Paralogous Proteins
Abstract
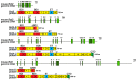
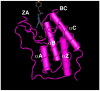
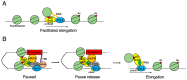
The Bromodomain and Extra-Terminal Domain (BET) family of proteins is characterized by the presence of two tandem bromodomains and an extra-terminal domain. The mammalian BET family of proteins comprises BRD2, BRD3, BRD4, and BRDT, which are encoded by paralogous genes that may have been generated by repeated duplication of an ancestral gene during evolution. Bromodomains that can specifically bind acetylated lysine residues in histones serve as chromatin-_targeting modules that decipher the histone acetylation code. BET proteins play a crucial role in regulating gene transcription through epigenetic interactions between bromodomains and acetylated histones during cellular proliferation and differentiation processes. On the other hand, BET proteins have been reported to mediate latent viral infection in host cells and be involved in oncogenesis. Human BRD4 is involved in multiple processes of the DNA virus life cycle, including viral replication, genome maintenance, and gene transcription through interaction with viral proteins. Aberrant BRD4 expression contributes to carcinogenesis by mediating hyperacetylation of the chromatin containing the cell proliferation-promoting genes. BET bromodomain blockade using small-molecule inhibitors gives rise to selective repression of the transcriptional network driven by c-MYC These inhibitors are expected to be potential therapeutic drugs for a wide range of cancers. This review presents an overview of the basic roles of BET proteins and highlights the pathological functions of BET and the recent developments in cancer therapy _targeting BET proteins in animal models.
Keywords: BET inhibitor; Bromodomain and Extra-Terminal Domain (BET); bromodomain; gene transcription; histone acetylation.
Conflict of interest statement
The author declares no conflict of interest.
Figures

Similar articles
-
BET Bromodomain as a _target of Epigenetic Therapy.Chem Pharm Bull (Tokyo). 2016;64(6):540-7. doi: 10.1248/cpb.c16-00225. Chem Pharm Bull (Tokyo). 2016. PMID: 27250788 Review.
-
Inhibition of BET bromodomains as a therapeutic strategy for cancer drug discovery.Onco_target. 2015 Mar 20;6(8):5501-16. doi: 10.18632/onco_target.3551. Onco_target. 2015. PMID: 25849938 Free PMC article. Review.
-
Small-Molecule _targeting of BET Proteins in Cancer.Adv Cancer Res. 2016;131:21-58. doi: 10.1016/bs.acr.2016.04.001. Epub 2016 May 31. Adv Cancer Res. 2016. PMID: 27451123 Review.
-
Bromodomains: Structure, function and pharmacology of inhibition.Biochem Pharmacol. 2016 Apr 15;106:1-18. doi: 10.1016/j.bcp.2015.12.005. Epub 2015 Dec 18. Biochem Pharmacol. 2016. PMID: 26707800 Review.
-
Epigenetic Readers of Lysine Acetylation Regulate Cocaine-Induced Plasticity.J Neurosci. 2015 Nov 11;35(45):15062-72. doi: 10.1523/JNEUROSCI.0826-15.2015. J Neurosci. 2015. PMID: 26558777 Free PMC article.
Cited by
-
Simultaneous administration of EZH2 and BET inhibitors inhibits proliferation and clonogenic ability of metastatic prostate cancer cells.J Enzyme Inhib Med Chem. 2023 Dec;38(1):2163242. doi: 10.1080/14756366.2022.2163242. J Enzyme Inhib Med Chem. 2023. PMID: 36629431 Free PMC article.
-
Bromodomain and Extraterminal Inhibition by JQ1 Produces Divergent Transcriptional Regulation of Suppressors of Cytokine Signaling Genes in Adipocytes.Endocrinology. 2020 Feb 1;161(2):bqz034. doi: 10.1210/endocr/bqz034. Endocrinology. 2020. PMID: 31875887 Free PMC article.
-
Epigenetics, DNA Organization, and Inflammatory Bowel Disease.Inflamm Bowel Dis. 2019 Jan 10;25(2):235-247. doi: 10.1093/ibd/izy330. Inflamm Bowel Dis. 2019. PMID: 30407525 Free PMC article. Review.
-
Epigenetic Reader Bromodomain Containing Protein 2 Facilitates Pathological Cardiac Hypertrophy via Regulating the Expression of Citrate Cycle Genes.Front Pharmacol. 2022 May 25;13:887991. doi: 10.3389/fphar.2022.887991. eCollection 2022. Front Pharmacol. 2022. PMID: 35694272 Free PMC article.
-
Bromodomain and Extra-Terminal Proteins in Brain Physiology and Pathology: BET-ing on Epigenetic Regulation.Biomedicines. 2023 Mar 1;11(3):750. doi: 10.3390/biomedicines11030750. Biomedicines. 2023. PMID: 36979729 Free PMC article. Review.
References
-
- Lygerou Z., Conesa C., Lesage P., Swanson R.N., Ruet A., Carlson M., Sentenac A., Seraphin B. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 1994;22:5332–5340. doi: 10.1093/nar/22.24.5332. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

