Analysis of intrapatient heterogeneity uncovers the microevolution of Middle East respiratory syndrome coronavirus
- PMID: 27900364
- PMCID: PMC5111008
- DOI: 10.1101/mcs.a001214
Analysis of intrapatient heterogeneity uncovers the microevolution of Middle East respiratory syndrome coronavirus
Abstract
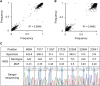
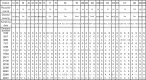
Genome sequence analysis of Middle East respiratory syndrome coronavirus (MERS-CoV) variants from patient specimens has revealed the evolutionary dynamics and mechanisms of pathogenesis of the virus. However, most studies have analyzed the consensus sequences of MERS-CoVs, precluding an investigation of intrapatient heterogeneity. Here, we analyzed non-consensus sequences to characterize intrapatient heterogeneity in cases associated with the 2015 outbreak of MERS in South Korea. Deep-sequencing analysis of MERS-CoV genomes performed on specimens from eight patients revealed significant intrapatient variation; therefore, sequence heterogeneity was further analyzed using _targeted deep sequencing. A total of 35 specimens from 24 patients (including a super-spreader) were sequenced to detect and analyze variants displaying intrapatient heterogeneity. Based on the analysis of non-consensus sequences, we demonstrated the intrapatient heterogeneity of MERS-CoVs, with the highest level in the super-spreader specimen. The heterogeneity could be transmitted in a close association with variation in the consensus sequences, suggesting the occurrence of multiple MERS-CoV infections. Analysis of intrapatient heterogeneity revealed a relationship between D510G and I529T mutations in the receptor-binding domain (RBD) of the viral spike glycoprotein. These two mutations have been reported to reduce the affinity of the RBD for human CD26. Notably, although the frequency of both D510G and I529T varied greatly among specimens, the combined frequency of the single mutants was consistently high (87.7% ± 1.9% on average). Concurrently, the frequency of occurrence of the wild type at the two positions was only 6.5% ± 1.7% on average, supporting the hypothesis that selection pressure exerted by the host immune response played a critical role in shaping genetic variants and their interaction in human MERS-CoVs during the outbreak.
Keywords: recurrent upper and lower respiratory tract infections.
Figures
Similar articles
-
Spread of Mutant Middle East Respiratory Syndrome Coronavirus with Reduced Affinity to Human CD26 during the South Korean Outbreak.mBio. 2016 Mar 1;7(2):e00019. doi: 10.1128/mBio.00019-16. mBio. 2016. PMID: 26933050 Free PMC article.
-
Mutations in the Spike Protein of Middle East Respiratory Syndrome Coronavirus Transmitted in Korea Increase Resistance to Antibody-Mediated Neutralization.J Virol. 2019 Jan 4;93(2):e01381-18. doi: 10.1128/JVI.01381-18. Print 2019 Jan 15. J Virol. 2019. PMID: 30404801 Free PMC article.
-
Recombinant Receptor-Binding Domains of Multiple Middle East Respiratory Syndrome Coronaviruses (MERS-CoVs) Induce Cross-Neutralizing Antibodies against Divergent Human and Camel MERS-CoVs and Antibody Escape Mutants.J Virol. 2016 Dec 16;91(1):e01651-16. doi: 10.1128/JVI.01651-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795425 Free PMC article.
-
Advances in MERS-CoV Vaccines and Therapeutics Based on the Receptor-Binding Domain.Viruses. 2019 Jan 14;11(1):60. doi: 10.3390/v11010060. Viruses. 2019. PMID: 30646569 Free PMC article. Review.
-
Korean Society for Laboratory Medicine Practice Guidelines for the Molecular Diagnosis of Middle East Respiratory Syndrome During an Outbreak in Korea in 2015.Ann Lab Med. 2016 May;36(3):203-8. doi: 10.3343/alm.2016.36.3.203. Ann Lab Med. 2016. PMID: 26915607 Free PMC article. Review.
Cited by
-
Why COVID-19 Transmission Is More Efficient and Aggressive Than Viral Transmission in Previous Coronavirus Epidemics?Biomolecules. 2020 Sep 11;10(9):1312. doi: 10.3390/biom10091312. Biomolecules. 2020. PMID: 32933047 Free PMC article. Review.
-
SARS-CoV-2 Point Mutation and Deletion Spectra and Their Association with Different Disease Outcomes.Microbiol Spectr. 2022 Apr 27;10(2):e0022122. doi: 10.1128/spectrum.00221-22. Epub 2022 Mar 29. Microbiol Spectr. 2022. PMID: 35348367 Free PMC article.
-
SARS-CoV-2 Quasispecies Provides an Advantage Mutation Pool for the Epidemic Variants.Microbiol Spectr. 2021 Sep 3;9(1):e0026121. doi: 10.1128/Spectrum.00261-21. Epub 2021 Aug 4. Microbiol Spectr. 2021. PMID: 34346744 Free PMC article.
-
Jumping a Moving Train: SARS-CoV-2 Evolution in Real Time.J Pediatric Infect Dis Soc. 2021 Dec 24;10(Supplement_4):S96-S105. doi: 10.1093/jpids/piab051. J Pediatric Infect Dis Soc. 2021. PMID: 34370041 Free PMC article. Review.
-
Current Status of Putative Animal Sources of SARS-CoV-2 Infection in Humans: Wildlife, Domestic Animals and Pets.Microorganisms. 2021 Apr 17;9(4):868. doi: 10.3390/microorganisms9040868. Microorganisms. 2021. PMID: 33920724 Free PMC article. Review.
References
-
- Briese T, Mishra N, Jain K, Zalmout IS, Jabado OJ, Karesh WB, Daszak P, Mohammed OB, Alagaili AN, Lipkin WI. 2014. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. MBio 5: e01146-14. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
