Effect of Dynamic Interaction between microRNA and Transcription Factor on Gene Expression
- PMID: 27957492
- PMCID: PMC5121577
- DOI: 10.1155/2016/2676282
Effect of Dynamic Interaction between microRNA and Transcription Factor on Gene Expression
Abstract
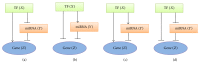
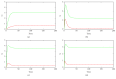
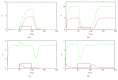
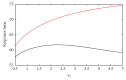
MicroRNAs (miRNAs) are endogenous noncoding RNAs which participate in diverse biological processes in animals and plants. They are known to join together with transcription factors and downstream gene, forming a complex and highly interconnected regulatory network. To recognize a few overrepresented motifs which are expected to perform important elementary regulatory functions, we constructed a computational model of miRNA-mediated feedforward loops (FFLs) in which a transcription factor (TF) regulates miRNA and _targets gene. Based on the different dynamic interactions between miRNA and TF on gene expression, four possible structural topologies of FFLs with two gate functions (AND gate and OR gate) are introduced. We studied the dynamic behaviors of these different motifs. Furthermore, the relationship between the response time and maximal activation velocity of miRNA was investigated. We found that the curve of response time shows nonmonotonic behavior in Co1 loop with OR gate. This may help us to infer the mechanism of miRNA binding to the promoter region. At last we investigated the influence of important parameters on the dynamic response of system. We identified that the stationary levels of _target gene in all loops were insensitive to the initial value of miRNA.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
Reconstruction and analysis of transcription factor-miRNA co-regulatory feed-forward loops in human cancers using filter-wrapper feature selection.PLoS One. 2013 Oct 29;8(10):e78197. doi: 10.1371/journal.pone.0078197. eCollection 2013. PLoS One. 2013. PMID: 24205155 Free PMC article.
-
A novel microRNA and transcription factor mediated regulatory network in schizophrenia.BMC Syst Biol. 2010 Feb 15;4:10. doi: 10.1186/1752-0509-4-10. BMC Syst Biol. 2010. PMID: 20156358 Free PMC article.
-
The role of incoherent microRNA-mediated feedforward loops in noise buffering.PLoS Comput Biol. 2011 Mar;7(3):e1001101. doi: 10.1371/journal.pcbi.1001101. Epub 2011 Mar 10. PLoS Comput Biol. 2011. PMID: 21423718 Free PMC article.
-
Transcription factor and microRNA co-regulatory loops: important regulatory motifs in biological processes and diseases.Brief Bioinform. 2015 Jan;16(1):45-58. doi: 10.1093/bib/bbt085. Epub 2013 Dec 4. Brief Bioinform. 2015. PMID: 24307685 Review.
-
Modeling microRNA-transcription factor networks in cancer.Adv Exp Med Biol. 2013;774:149-67. doi: 10.1007/978-94-007-5590-1_9. Adv Exp Med Biol. 2013. PMID: 23377973 Review.
Cited by
-
Identification of MicroRNA-_target Gene-Transcription Factor Regulatory Networks in Colorectal Adenoma Using Microarray Expression Data.Front Genet. 2020 May 19;11:463. doi: 10.3389/fgene.2020.00463. eCollection 2020. Front Genet. 2020. PMID: 32508878 Free PMC article.
-
Deciphering the Molecular Mechanisms of Autonomic Nervous System Neuron Induction through Integrative Bioinformatics Analysis.Int J Mol Sci. 2023 May 21;24(10):9053. doi: 10.3390/ijms24109053. Int J Mol Sci. 2023. PMID: 37240399 Free PMC article.
-
From Endogenous to Synthetic microRNA-Mediated Regulatory Circuits: An Overview.Cells. 2019 Nov 29;8(12):1540. doi: 10.3390/cells8121540. Cells. 2019. PMID: 31795372 Free PMC article. Review.
-
Integrated analysis of multiple bioinformatics studies to identify microRNA-_target gene-transcription factor regulatory networks in retinoblastoma.Transl Cancer Res. 2022 Jul;11(7):2225-2237. doi: 10.21037/tcr-21-1748. Transl Cancer Res. 2022. PMID: 35966326 Free PMC article.
-
Identification of Differentially Expressed Genes and Prediction of Expression Regulation Networks in Dysfunctional Endothelium.Genes (Basel). 2022 Aug 30;13(9):1563. doi: 10.3390/genes13091563. Genes (Basel). 2022. PMID: 36140731 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

