Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor
- PMID: 28017329
- PMCID: PMC5235968
- DOI: 10.1016/j.cell.2016.11.042
Prohibitin 2 Is an Inner Mitochondrial Membrane Mitophagy Receptor
Abstract
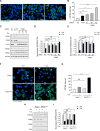
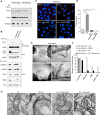
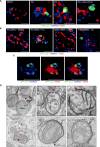
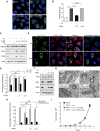
The removal of unwanted or damaged mitochondria by autophagy, a process called mitophagy, is essential for key events in development, cellular homeostasis, tumor suppression, and prevention of neurodegeneration and aging. However, the precise mechanisms of mitophagy remain uncertain. Here, we identify the inner mitochondrial membrane protein, prohibitin 2 (PHB2), as a crucial mitophagy receptor involved in _targeting mitochondria for autophagic degradation. PHB2 binds the autophagosomal membrane-associated protein LC3 through an LC3-interaction region (LIR) domain upon mitochondrial depolarization and proteasome-dependent outer membrane rupture. PHB2 is required for Parkin-induced mitophagy in mammalian cells and for the clearance of paternal mitochondria after embryonic fertilization in C. elegans. Our findings pinpoint a conserved mechanism of eukaryotic mitophagy and demonstrate a function of prohibitin 2 that may underlie its roles in physiology, aging, and disease.
Keywords: autophagy; mitophagy; prohibitin 2.
Copyright © 2017 Elsevier Inc. All rights reserved.
Figures


Comment in
-
PHB2/prohibitin 2: An inner membrane mitophagy receptor.Cell Res. 2017 Mar;27(3):311-312. doi: 10.1038/cr.2017.23. Epub 2017 Feb 21. Cell Res. 2017. PMID: 28220775 Free PMC article.
Similar articles
-
PHB2 (prohibitin 2) promotes PINK1-PRKN/Parkin-dependent mitophagy by the PARL-PGAM5-PINK1 axis.Autophagy. 2020 Mar;16(3):419-434. doi: 10.1080/15548627.2019.1628520. Epub 2019 Jun 16. Autophagy. 2020. PMID: 31177901 Free PMC article.
-
Dimerization of mitophagy receptor BNIP3L/NIX is essential for recruitment of autophagic machinery.Autophagy. 2021 May;17(5):1232-1243. doi: 10.1080/15548627.2020.1755120. Epub 2020 Apr 24. Autophagy. 2021. PMID: 32286918 Free PMC article.
-
Fndc-1 contributes to paternal mitochondria elimination in C. elegans.Dev Biol. 2019 Oct 1;454(1):15-20. doi: 10.1016/j.ydbio.2019.06.016. Epub 2019 Jun 21. Dev Biol. 2019. PMID: 31233739 Free PMC article.
-
Mitophagy during development and stress in C. elegans.Mech Ageing Dev. 2020 Jul;189:111266. doi: 10.1016/j.mad.2020.111266. Epub 2020 May 23. Mech Ageing Dev. 2020. PMID: 32454052 Review.
-
Autophagosomal Sperm Organelle Clearance and mtDNA Inheritance in C. elegans.Adv Anat Embryol Cell Biol. 2019;231:1-23. doi: 10.1007/102_2018_1. Adv Anat Embryol Cell Biol. 2019. PMID: 30467692 Review.
Cited by
-
PHB2 promotes SHIP2 ubiquitination via the E3 ligase NEDD4 to regulate AKT signaling in gastric cancer.J Exp Clin Cancer Res. 2024 Jan 11;43(1):17. doi: 10.1186/s13046-023-02937-1. J Exp Clin Cancer Res. 2024. PMID: 38200519 Free PMC article.
-
ATAD3B is a mitophagy receptor mediating clearance of oxidative stress-induced damaged mitochondrial DNA.EMBO J. 2021 Apr 15;40(8):e106283. doi: 10.15252/embj.2020106283. Epub 2021 Mar 5. EMBO J. 2021. PMID: 33665835 Free PMC article.
-
Nitrosative Redox Homeostasis and Antioxidant Response Defense in Disused Vastus lateralis Muscle in Long-Term Bedrest (Toulouse Cocktail Study).Antioxidants (Basel). 2021 Mar 3;10(3):378. doi: 10.3390/antiox10030378. Antioxidants (Basel). 2021. PMID: 33802593 Free PMC article.
-
The connection between the dynamic remodeling of the mitochondrial network and the regulation of muscle mass.Cell Mol Life Sci. 2021 Feb;78(4):1305-1328. doi: 10.1007/s00018-020-03662-0. Epub 2020 Oct 19. Cell Mol Life Sci. 2021. PMID: 33078210 Free PMC article. Review.
-
A novel mechanism of PHB2-mediated mitophagy participating in the development of Parkinson's disease.Neural Regen Res. 2024 Aug 1;19(8):1828-1834. doi: 10.4103/1673-5374.389356. Epub 2023 Nov 8. Neural Regen Res. 2024. PMID: 38103250 Free PMC article.
References
-
- Al Rawi S, Louvet-Vallée S, Djeddi A, Sachse M, Culetto E, Hajjar C, Boyd L, Legouis R, Galy V. Postfertilization autophagy of sperm organelles prevents paternal mitochondrial DNA transmission. Science. 2011;334:1144–1147. - PubMed
-
- Artal-Sanz M, Tavernarakis N. Prohibitin and mitochondrial biology. Trends Endocrinol Metab. 2009;20:394–401. - PubMed
-
- Bingol B, Sheng M. Mechanisms of mitophagy: PINK1, Parkin, USP30 and beyond. Free Radic Biol Med. 2016;S0891:30033–30038. - PubMed
-
- Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature. 2014;510:370–375. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

