Long noncoding RNA repertoire in chicken liver and adipose tissue
- PMID: 28073357
- PMCID: PMC5225574
- DOI: 10.1186/s12711-016-0275-0
Long noncoding RNA repertoire in chicken liver and adipose tissue
Abstract
Background: Improving functional annotation of the chicken genome is a key challenge in bridging the gap between genotype and phenotype. Among all transcribed regions, long noncoding RNAs (lncRNAs) are a major component of the transcriptome and its regulation, and whole-transcriptome sequencing (RNA-Seq) has greatly improved their identification and characterization. We performed an extensive profiling of the lncRNA transcriptome in the chicken liver and adipose tissue by RNA-Seq. We focused on these two tissues because of their importance in various economical traits for which energy storage and mobilization play key roles and also because of their high cell homogeneity. To predict lncRNAs, we used a recently developed tool called FEELnc, which also classifies them with respect to their distance and strand orientation to the closest protein-coding genes. Moreover, to confidently identify the genes/transcripts expressed in each tissue (a complex task for weakly expressed molecules such as lncRNAs), we probed a particularly large number of biological replicates (16 per tissue) compared to common multi-tissue studies with a larger set of tissues but less sampling.
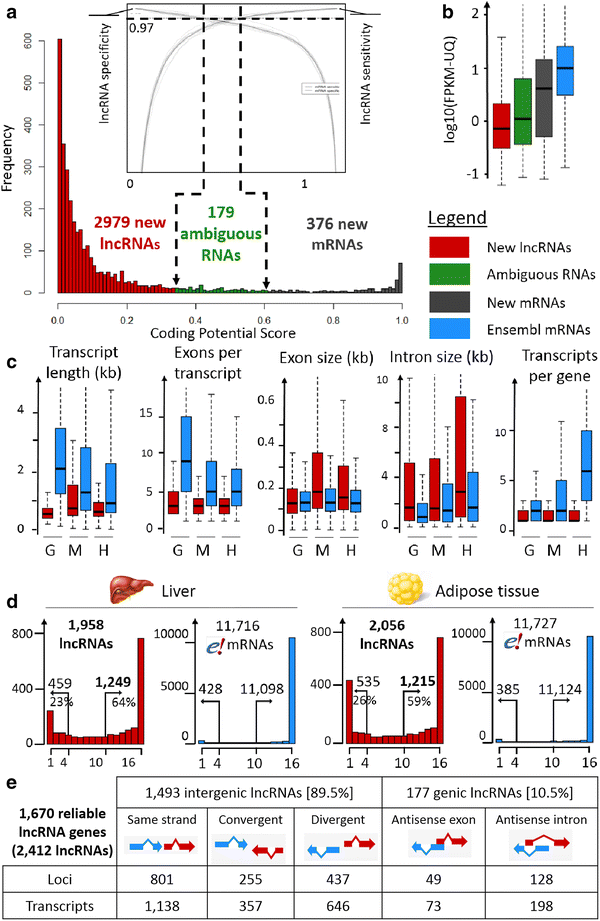
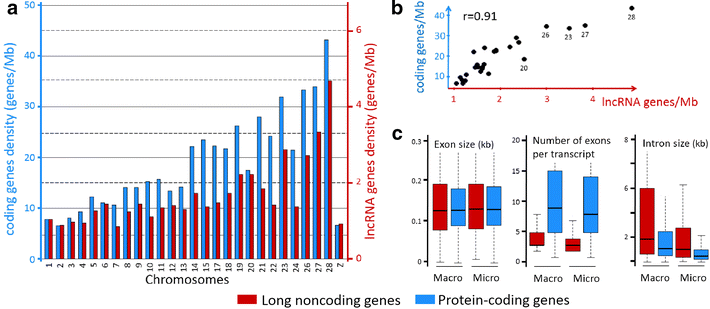
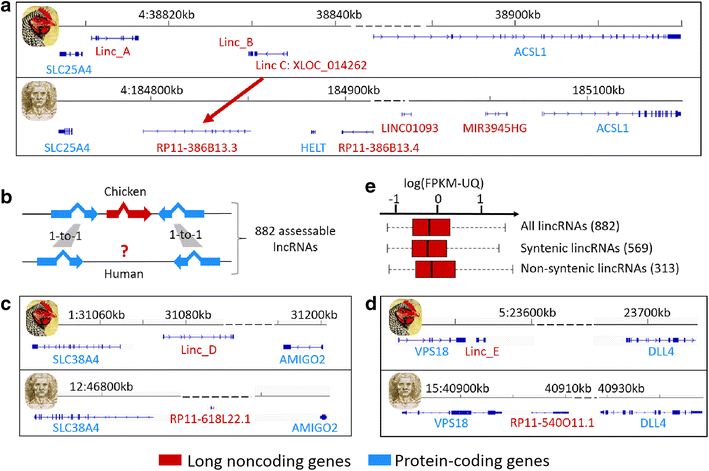
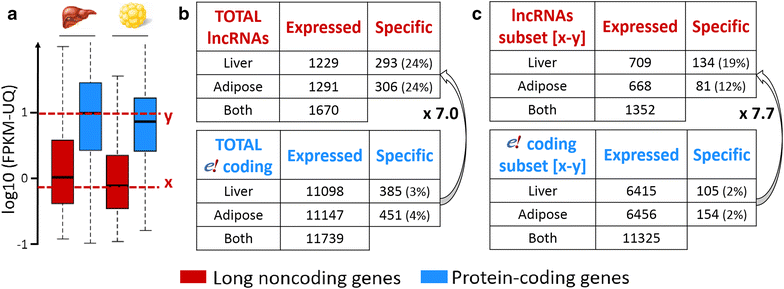
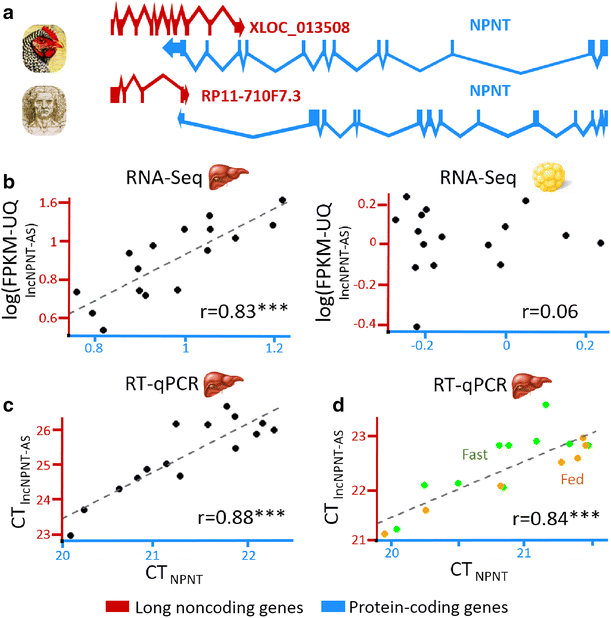
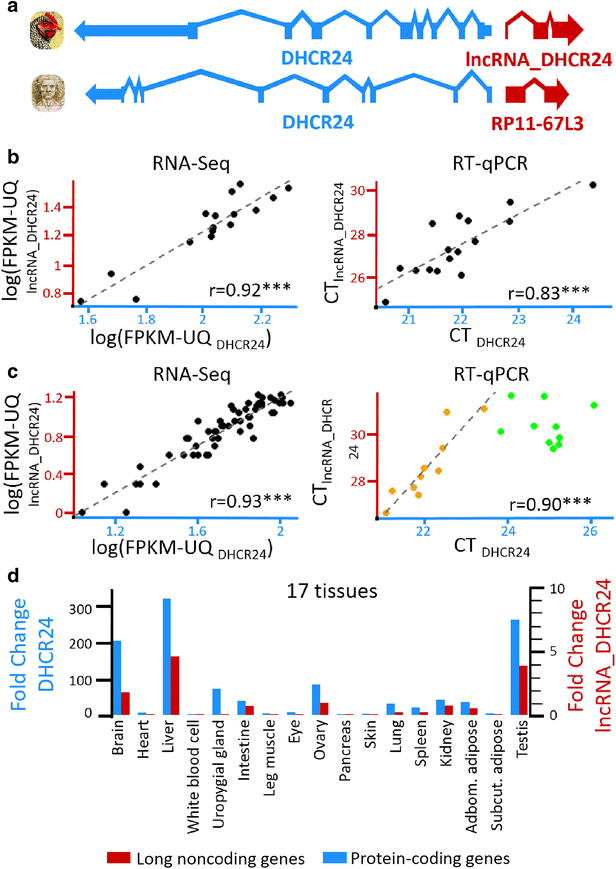
Results: We predicted 2193 lncRNA genes, among which 1670 were robustly expressed across replicates in the liver and/or adipose tissue and which were classified into 1493 intergenic and 177 intragenic lncRNAs located between and within protein-coding genes, respectively. We observed similar structural features between chickens and mammals, with strong synteny conservation but without sequence conservation. As previously reported, we confirm that lncRNAs have a lower and more tissue-specific expression than mRNAs. Finally, we showed that adjacent lncRNA-mRNA genes in divergent orientation have a higher co-expression level when separated by less than 1 kb compared to more distant divergent pairs. Among these, we highlighted for the first time a novel lncRNA candidate involved in lipid metabolism, lnc_DHCR24, which is highly correlated with the DHCR24 gene that encodes a key enzyme of cholesterol biosynthesis.
Conclusions: We provide a comprehensive lncRNA repertoire in the chicken liver and adipose tissue, which shows interesting patterns of co-expression between mRNAs and lncRNAs. It contributes to improving the structural and functional annotation of the chicken genome and provides a basis for further studies on energy storage and mobilization traits in the chicken.
Figures
Similar articles
-
Transcriptome Profile Analysis Reveals an Estrogen Induced LncRNA Associated with Lipid Metabolism and Carcass Traits in Chickens (Gallus Gallus).Cell Physiol Biochem. 2018;50(5):1638-1658. doi: 10.1159/000494785. Epub 2018 Nov 1. Cell Physiol Biochem. 2018. PMID: 30384372
-
Genome-wide identification of tissue-specific long non-coding RNA in three farm animal species.BMC Genomics. 2018 Sep 18;19(1):684. doi: 10.1186/s12864-018-5037-7. BMC Genomics. 2018. PMID: 30227846 Free PMC article.
-
Analyses of Long Non-Coding RNA and mRNA profiling using RNA sequencing in chicken testis with extreme sperm motility.Sci Rep. 2017 Aug 22;7(1):9055. doi: 10.1038/s41598-017-08738-9. Sci Rep. 2017. PMID: 28831078 Free PMC article.
-
Long noncoding RNAs in lipid metabolism: literature review and conservation analysis across species.BMC Genomics. 2019 Nov 21;20(1):882. doi: 10.1186/s12864-019-6093-3. BMC Genomics. 2019. PMID: 31752679 Free PMC article. Review.
-
The specificity of long noncoding RNA expression.Biochim Biophys Acta. 2016 Jan;1859(1):16-22. doi: 10.1016/j.bbagrm.2015.08.005. Epub 2015 Aug 19. Biochim Biophys Acta. 2016. PMID: 26297315 Review.
Cited by
-
Multi-species annotation of transcriptome and chromatin structure in domesticated animals.BMC Biol. 2019 Dec 30;17(1):108. doi: 10.1186/s12915-019-0726-5. BMC Biol. 2019. PMID: 31884969 Free PMC article.
-
Transcriptomes of an Array of Chicken Ovary, Intestinal, and Immune Cells and Tissues.Front Genet. 2021 Jun 30;12:664424. doi: 10.3389/fgene.2021.664424. eCollection 2021. Front Genet. 2021. PMID: 34276773 Free PMC article.
-
Genome-wide identification and characterization of long non-coding RNAs during postnatal development of rabbit adipose tissue.Lipids Health Dis. 2018 Nov 28;17(1):271. doi: 10.1186/s12944-018-0915-1. Lipids Health Dis. 2018. PMID: 30486837 Free PMC article.
-
Landscape of long non-coding RNAs in Trichophyton mentagrophytes-induced rabbit dermatophytosis lesional skin and normal skin.Funct Integr Genomics. 2018 Jul;18(4):401-410. doi: 10.1007/s10142-018-0601-4. Epub 2018 Mar 20. Funct Integr Genomics. 2018. PMID: 29560532
-
Multi-tissue observation of the long non-coding RNA effects on sexually biased gene expression in cattle.Asian-Australas J Anim Sci. 2019 Jul;32(7):1044-1051. doi: 10.5713/ajas.18.0516. Epub 2018 Nov 28. Asian-Australas J Anim Sci. 2019. PMID: 30744377 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

