xMAP Technology: Applications in Detection of Pathogens
- PMID: 28179899
- PMCID: PMC5263158
- DOI: 10.3389/fmicb.2017.00055
xMAP Technology: Applications in Detection of Pathogens
Abstract
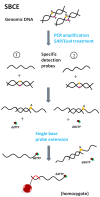
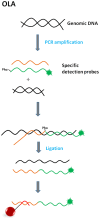
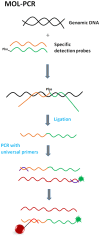
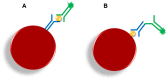
xMAP technology is applicable for high-throughput, multiplex and simultaneous detection of different analytes within a single complex sample. xMAP multiplex assays are currently available in various nucleic acid and immunoassay formats, enabling simultaneous detection and typing of pathogenic viruses, bacteria, parasites and fungi and also antigen or antibody interception. As an open architecture platform, the xMAP technology is beneficial to end users and therefore it is used in various pharmaceutical, clinical and research laboratories. The main aim of this review is to summarize the latest findings and applications in the field of pathogen detection using microsphere-based multiplex assays.
Keywords: diagnostics; immunoassay; magnetic microspheres; multiplex detection; nucleic acid detection; pathogen identification; xMAP.
Figures

Similar articles
-
Conversion of a capture ELISA to a Luminex xMAP assay using a multiplex antibody screening method.J Vis Exp. 2012 Jul 6;(65):4084. doi: 10.3791/4084. J Vis Exp. 2012. PMID: 22806215 Free PMC article.
-
The genesis and evolution of bead-based multiplexing.Methods. 2019 Apr 1;158:2-11. doi: 10.1016/j.ymeth.2019.01.007. Epub 2019 Jan 17. Methods. 2019. PMID: 30659874 Review.
-
Opportunities for bead-based multiplex assays in veterinary diagnostic laboratories.J Vet Diagn Invest. 2013 Nov;25(6):671-91. doi: 10.1177/1040638713507256. Epub 2013 Oct 23. J Vet Diagn Invest. 2013. PMID: 24153036 Review.
-
Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection.Clin Chim Acta. 2006 Jan;363(1-2):71-82. doi: 10.1016/j.cccn.2005.06.023. Epub 2005 Aug 15. Clin Chim Acta. 2006. PMID: 16102740 Free PMC article. Review.
-
[A rapid, multiplexed new technology xMAP liquid chip for detection and identification of pathogens].Wei Sheng Yan Jiu. 2007 Nov;36(6):759-62. Wei Sheng Yan Jiu. 2007. PMID: 18303645 Review. Chinese.
Cited by
-
Quantitative assessment of multiple pathogen exposure and immune dynamics at scale.Microbiol Spectr. 2024 Jan 11;12(1):e0239923. doi: 10.1128/spectrum.02399-23. Epub 2023 Dec 8. Microbiol Spectr. 2024. PMID: 38063388 Free PMC article.
-
Modernization of Control of Pathogenic Micro-Organisms in the Food-Chain Requires a Durable Role for Immunoaffinity-Based Detection Methodology-A Review.Foods. 2021 Apr 11;10(4):832. doi: 10.3390/foods10040832. Foods. 2021. PMID: 33920486 Free PMC article. Review.
-
Isotopically Encoded Nanotags for Multiplexed Ion Beam Imaging.Adv Mater Technol. 2020 Jul;5(7):2000098. doi: 10.1002/admt.202000098. Epub 2020 May 6. Adv Mater Technol. 2020. PMID: 32661501 Free PMC article.
-
Liquid Biopsies, Novel Approaches and Future Directions.Cancers (Basel). 2023 Mar 3;15(5):1579. doi: 10.3390/cancers15051579. Cancers (Basel). 2023. PMID: 36900369 Free PMC article. Review.
-
Simultaneous Immunodetection of Anthrax, Plague, and Tularemia from Blood Cultures by Use of Multiplexed Suspension Arrays.J Clin Microbiol. 2018 Mar 26;56(4):e01479-17. doi: 10.1128/JCM.01479-17. Print 2018 Apr. J Clin Microbiol. 2018. PMID: 29386263 Free PMC article.
References
-
- Anderson J. P., Rascoe L. N., Levert K., Chastain H. M., Reed M. S., Rivera H. N., et al. (2015). Development of a luminex bead based assay for diagnosis of toxocariasis using recombinant antigens Tc-CTL-1 and Tc-TES-26. PLoS Negl. Trop. Dis. 9:e0004168 10.1371/journal.pntd.0004168 - DOI - PMC - PubMed
-
- Angeloni S., Cordes R., Dunbar S., Garcia C., Gibson G., Martin C., et al. (2014). xMAP Cookbook: A Collection of Methods and Protocols for Developing Multiplex Assays with xMAP Technology, 2nd Edn (Austin, TX: Luminex; )
-
- Anonymous. (2014). Multicriteria-Based Ranking for Risk Management of Food-Borne Parasites: Microbiological Risk Assessment Series (MRA) 23. Rome: FAO, 324.
-
- Anonymous. (2015). Regulation (EU) 2015/1375, laying down specific rules on official controls for Trichinella in meat. Official J. Eur. Union 28 7.
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

