ERRα induces H3K9 demethylation by LSD1 to promote cell invasion
- PMID: 28348226
- PMCID: PMC5393192
- DOI: 10.1073/pnas.1614664114
ERRα induces H3K9 demethylation by LSD1 to promote cell invasion
Abstract
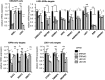
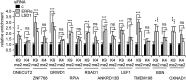
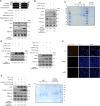
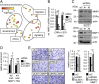
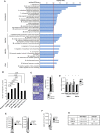
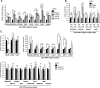
Lysine Specific Demethylase 1 (LSD1) removes mono- and dimethyl groups from lysine 4 of histone H3 (H3K4) or H3K9, resulting in repressive or activating (respectively) transcriptional histone marks. The mechanisms that control the balance between these two antagonist activities are not understood. We here show that LSD1 and the orphan nuclear receptor estrogen-related receptor α (ERRα) display commonly activated genes. Transcriptional activation by LSD1 and ERRα involves H3K9 demethylation at the transcriptional start site (TSS). Strikingly, ERRα is sufficient to induce LSD1 to demethylate H3K9 in vitro. The relevance of this mechanism is highlighted by functional data. LSD1 and ERRα coregulate several _target genes involved in cell migration, including the MMP1 matrix metallo-protease, also activated through H3K9 demethylation at the TSS. Depletion of LSD1 or ERRα reduces the cellular capacity to invade the extracellular matrix, a phenomenon that is rescued by MMP1 reexpression. Altogether our results identify a regulatory network involving a direct switch in the biochemical activities of a histone demethylase, leading to increased cell invasion.
Keywords: ERRα; LSD1; cell migration; histone demethylation; transcriptional regulation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
ERRα protein is stabilized by LSD1 in a demethylation-independent manner.PLoS One. 2017 Nov 30;12(11):e0188871. doi: 10.1371/journal.pone.0188871. eCollection 2017. PLoS One. 2017. PMID: 29190800 Free PMC article.
-
LSD1-ERRα complex requires NRF1 to positively regulate transcription and cell invasion.Sci Rep. 2018 Jul 3;8(1):10041. doi: 10.1038/s41598-018-27676-8. Sci Rep. 2018. PMID: 29968728 Free PMC article.
-
Lysine-specific demethylase 1-mediated demethylation of histone H3 lysine 9 contributes to interleukin 1β-induced microsomal prostaglandin E synthase 1 expression in human osteoarthritic chondrocytes.Arthritis Res Ther. 2014 May 16;16(3):R113. doi: 10.1186/ar4564. Arthritis Res Ther. 2014. PMID: 24886859 Free PMC article.
-
LSD1: more than demethylation of histone lysine residues.Exp Mol Med. 2020 Dec;52(12):1936-1947. doi: 10.1038/s12276-020-00542-2. Epub 2020 Dec 14. Exp Mol Med. 2020. PMID: 33318631 Free PMC article. Review.
-
Post-translational modifications of lysine-specific demethylase 1.Biochim Biophys Acta Gene Regul Mech. 2023 Dec;1866(4):194968. doi: 10.1016/j.bbagrm.2023.194968. Epub 2023 Aug 10. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 37572976 Review.
Cited by
-
The PRMT5-LSD1 axis confers Slug dual transcriptional activities and promotes breast cancer progression.J Exp Clin Cancer Res. 2022 Jun 2;41(1):191. doi: 10.1186/s13046-022-02400-7. J Exp Clin Cancer Res. 2022. PMID: 35655230 Free PMC article.
-
Repression of LSD1 potentiates homologous recombination-proficient ovarian cancer to PARP inhibitors through down-regulation of BRCA1/2 and RAD51.Nat Commun. 2023 Nov 16;14(1):7430. doi: 10.1038/s41467-023-42850-x. Nat Commun. 2023. PMID: 37973845 Free PMC article.
-
A functional LSD1 coregulator screen reveals a novel transcriptional regulatory cascade connecting R-loop homeostasis with epigenetic regulation.Nucleic Acids Res. 2021 May 7;49(8):4350-4370. doi: 10.1093/nar/gkab180. Nucleic Acids Res. 2021. PMID: 33823549 Free PMC article.
-
ERRα: unraveling its role as a key player in cell migration.Oncogene. 2024 Feb;43(6):379-387. doi: 10.1038/s41388-023-02899-w. Epub 2023 Dec 21. Oncogene. 2024. PMID: 38129506 Review.
-
ERRα protein is stabilized by LSD1 in a demethylation-independent manner.PLoS One. 2017 Nov 30;12(11):e0188871. doi: 10.1371/journal.pone.0188871. eCollection 2017. PLoS One. 2017. PMID: 29190800 Free PMC article.
References
-
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128(4):707–719. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293(5532):1074–1080. - PubMed
-
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20(11):1405–1428. - PubMed
-
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128(4):693–705. - PubMed
-
- Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25(1):1–14. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

