Virus genomes reveal factors that spread and sustained the Ebola epidemic
- PMID: 28405027
- PMCID: PMC5712493
- DOI: 10.1038/nature22040
Virus genomes reveal factors that spread and sustained the Ebola epidemic
Abstract
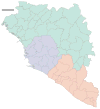
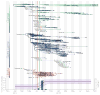
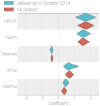
The 2013-2016 West African epidemic caused by the Ebola virus was of unprecedented magnitude, duration and impact. Here we reconstruct the dispersal, proliferation and decline of Ebola virus throughout the region by analysing 1,610 Ebola virus genomes, which represent over 5% of the known cases. We test the association of geography, climate and demography with viral movement among administrative regions, inferring a classic 'gravity' model, with intense dispersal between larger and closer populations. Despite attenuation of international dispersal after border closures, cross-border transmission had already sown the seeds for an international epidemic, rendering these measures ineffective at curbing the epidemic. We address why the epidemic did not spread into neighbouring countries, showing that these countries were susceptible to substantial outbreaks but at lower risk of introductions. Finally, we reveal that this large epidemic was a heterogeneous and spatially dissociated collection of transmission clusters of varying size, duration and connectivity. These insights will help to inform interventions in future epidemics.
Conflict of interest statement
The authors declare no competing financial interests.
Figures




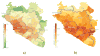
Similar articles
-
Phylodynamic assessment of intervention strategies for the West African Ebola virus outbreak.Nat Commun. 2018 Jun 8;9(1):2222. doi: 10.1038/s41467-018-03763-2. Nat Commun. 2018. PMID: 29884821 Free PMC article.
-
The evolution of Ebola virus: Insights from the 2013-2016 epidemic.Nature. 2016 Oct 13;538(7624):193-200. doi: 10.1038/nature19790. Nature. 2016. PMID: 27734858 Free PMC article. Review.
-
Genomic surveillance elucidates Ebola virus origin and transmission during the 2014 outbreak.Science. 2014 Sep 12;345(6202):1369-72. doi: 10.1126/science.1259657. Epub 2014 Aug 28. Science. 2014. PMID: 25214632 Free PMC article.
-
Ebola Virus Epidemiology, Transmission, and Evolution during Seven Months in Sierra Leone.Cell. 2015 Jun 18;161(7):1516-26. doi: 10.1016/j.cell.2015.06.007. Cell. 2015. PMID: 26091036 Free PMC article.
-
Learning from Ebola Virus: How to Prevent Future Epidemics.Viruses. 2015 Jul 9;7(7):3789-97. doi: 10.3390/v7072797. Viruses. 2015. PMID: 26184283 Free PMC article. Review.
Cited by
-
Modeling international mobility using roaming cell phone traces during COVID-19 pandemic.EPJ Data Sci. 2022;11(1):22. doi: 10.1140/epjds/s13688-022-00335-9. Epub 2022 Apr 4. EPJ Data Sci. 2022. PMID: 35402140 Free PMC article.
-
Phylogenetic and phylodynamic approaches to understanding and combating the early SARS-CoV-2 pandemic.Nat Rev Genet. 2022 Sep;23(9):547-562. doi: 10.1038/s41576-022-00483-8. Epub 2022 Apr 22. Nat Rev Genet. 2022. PMID: 35459859 Free PMC article. Review.
-
Phycova - a tool for exploring covariates of pathogen spread.Virus Evol. 2022 Feb 18;8(1):veac015. doi: 10.1093/ve/veac015. eCollection 2022. Virus Evol. 2022. PMID: 35295748 Free PMC article.
-
Outbreak analytics: a developing data science for informing the response to emerging pathogens.Philos Trans R Soc Lond B Biol Sci. 2019 Jul 8;374(1776):20180276. doi: 10.1098/rstb.2018.0276. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 31104603 Free PMC article. Review.
-
Capturing sequence diversity in metagenomes with comprehensive and scalable probe design.Nat Biotechnol. 2019 Feb;37(2):160-168. doi: 10.1038/s41587-018-0006-x. Epub 2019 Feb 4. Nat Biotechnol. 2019. PMID: 30718881 Free PMC article.
References
-
- World Health Organization. Ebola situation report - 10 june 2016. 2016 URL http://apps.who.int/iris/bitstream/10665/208883/1/ebolasitrep_10Jun2016_....
-
- Kuhn JH, et al. Nomenclature- and database-compatible names for the two ebola virus variants that emerged in guinea and the democratic republic of the congo in 2014. Viruses. 2014;6:4760–4799. URL http://dx.doi.org/10.3390/v6114760. - DOI - PMC - PubMed
-
- Baize S, et al. Emergence of zaire ebola virus disease in guinea. The New England Journal of Medicine. 2014;371:1418–1425. URL http://dx.doi.org/10.1056/NEJMoa1404505. - DOI - PubMed
-
- World Health Organization Regional Office for Africa. Ebola virus disease, west africa (situation as of 25 april 2014) 2014 URL http://www.afro.who.int/en/clusters-a-programmes/dpc/epidemic-a-pandemic....
-
- Goba A, et al. An outbreak of ebola virus disease in the lassa fever zone. The Journal of infectious diseases. 2016 URL http://dx.doi.org/10.1093/infdis/jiw239. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
- U01 AI082119/AI/NIAID NIH HHS/United States
- R01 AI114855/AI/NIAID NIH HHS/United States
- R01 AI081982/AI/NIAID NIH HHS/United States
- R01 AI107034/AI/NIAID NIH HHS/United States
- UL1 TR001114/TR/NCATS NIH HHS/United States
- UL1 RR025774/RR/NCRR NIH HHS/United States
- R35 GM119774/GM/NIGMS NIH HHS/United States
- 106866/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- 5R01AI/114855-03/National Institute of Allergy and Infectious Disease/International
- R01 AI104621/AI/NIAID NIH HHS/United States
- AI082119/NH/NIH HHS/United States
- AI081982/NH/NIH HHS/United States
- HHSN272201400048C/AI/NIAID NIH HHS/United States
- AI082805/NH/NIH HHS/United States
- MC_UU_12014/12/MRC_/Medical Research Council/United Kingdom
- R01 AI117011/AI/NIAID NIH HHS/United States
- R01 HG006139/HG/NHGRI NIH HHS/United States
- HHSN272200700016I/AI/NIAID NIH HHS/United States
- R13 AI104216/AI/NIAID NIH HHS/United States
- U01 HG007480/HG/NHGRI NIH HHS/United States
- AI104621/NH/NIH HHS/United States
- AI104216/NH/NIH HHS/United States
- R44 AI115754/AI/NIAID NIH HHS/United States
- AI115754/NH/NIH HHS/United States
- MR/L015080/1/MRC_/Medical Research Council/United Kingdom
- 260864/ERC_/European Research Council/International
- R44 AI088843/AI/NIAID NIH HHS/United States
- S10 OD020069/OD/NIH HHS/United States
- AI088843/NH/NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- 001/WHO_/World Health Organization/International
- MR/M501621/1/MRC_/Medical Research Council/United Kingdom
- R43 AI088843/AI/NIAID NIH HHS/United States
- 1U01HG007480-01/NH/NIH HHS/United States
- U19 AI110818/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

