Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations
- PMID: 28445466
- PMCID: PMC5427175
- DOI: 10.1038/nature22312
Human pluripotent stem cells recurrently acquire and expand dominant negative P53 mutations
Abstract
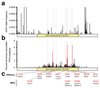
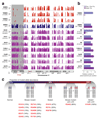
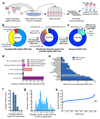
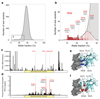
Human pluripotent stem cells (hPS cells) can self-renew indefinitely, making them an attractive source for regenerative therapies. This expansion potential has been linked with the acquisition of large copy number variants that provide mutated cells with a growth advantage in culture. The nature, extent and functional effects of other acquired genome sequence mutations in cultured hPS cells are not known. Here we sequence the protein-coding genes (exomes) of 140 independent human embryonic stem cell (hES cell) lines, including 26 lines prepared for potential clinical use. We then apply computational strategies for identifying mutations present in a subset of cells in each hES cell line. Although such mosaic mutations were generally rare, we identified five unrelated hES cell lines that carried six mutations in the TP53 gene that encodes the tumour suppressor P53. The TP53 mutations we observed are dominant negative and are the mutations most commonly seen in human cancers. We found that the TP53 mutant allelic fraction increased with passage number under standard culture conditions, suggesting that the P53 mutations confer selective advantage. We then mined published RNA sequencing data from 117 hPS cell lines, and observed another nine TP53 mutations, all resulting in coding changes in the DNA-binding domain of P53. In three lines, the allelic fraction exceeded 50%, suggesting additional selective advantage resulting from the loss of heterozygosity at the TP53 locus. As the acquisition and expansion of cancer-associated mutations in hPS cells may go unnoticed during most applications, we suggest that careful genetic characterization of hPS cells and their differentiated derivatives be carried out before clinical use.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


Comment in
-
Stem cells: Subclone wars.Nature. 2017 May 11;545(7653):160-161. doi: 10.1038/nature22490. Epub 2017 Apr 26. Nature. 2017. PMID: 28445463 No abstract available.
-
Stem cells: Stem cell-based therapies threatened by the accumulation of p53 mutations.Nat Rev Mol Cell Biol. 2017 Jul;18(7):403. doi: 10.1038/nrm.2017.52. Epub 2017 May 17. Nat Rev Mol Cell Biol. 2017. PMID: 28512352 No abstract available.
Similar articles
-
Spontaneous Single-Copy Loss of TP53 in Human Embryonic Stem Cells Markedly Increases Cell Proliferation and Survival.Stem Cells. 2017 Apr;35(4):872-885. doi: 10.1002/stem.2550. Epub 2017 Jan 19. Stem Cells. 2017. PMID: 27888558
-
High prevalence of acquired cancer-related mutations in 146 human pluripotent stem cell lines and their differentiated derivatives.Nat Biotechnol. 2024 Nov;42(11):1667-1671. doi: 10.1038/s41587-023-02090-2. Epub 2024 Jan 9. Nat Biotechnol. 2024. PMID: 38195986
-
Mutational processes shape the landscape of TP53 mutations in human cancer.Nat Genet. 2018 Oct;50(10):1381-1387. doi: 10.1038/s41588-018-0204-y. Epub 2018 Sep 17. Nat Genet. 2018. PMID: 30224644 Free PMC article.
-
Role of p53 in regulation of hematopoiesis in health and disease.Curr Opin Hematol. 2022 Jul 1;29(4):194-200. doi: 10.1097/MOH.0000000000000715. Epub 2022 Mar 7. Curr Opin Hematol. 2022. PMID: 35787548 Review.
-
TP53 mutations in cancer: Molecular features and therapeutic opportunities (Review).Int J Mol Med. 2025 Jan;55(1):7. doi: 10.3892/ijmm.2024.5448. Epub 2024 Oct 25. Int J Mol Med. 2025. PMID: 39450536 Free PMC article. Review.
Cited by
-
Efficient and safe single-cell cloning of human pluripotent stem cells using the CEPT cocktail.Nat Protoc. 2023 Jan;18(1):58-80. doi: 10.1038/s41596-022-00753-z. Epub 2022 Oct 19. Nat Protoc. 2023. PMID: 36261632 Free PMC article. Review.
-
Harnessing cellular therapeutics for type 1 diabetes mellitus: progress, challenges, and the road ahead.Nat Rev Endocrinol. 2025 Jan;21(1):14-30. doi: 10.1038/s41574-024-01029-0. Epub 2024 Sep 3. Nat Rev Endocrinol. 2025. PMID: 39227741 Review.
-
Large animal model species in pluripotent stem cell therapy research and development for retinal diseases: a systematic review.Front Ophthalmol (Lausanne). 2024 Aug 26;4:1377098. doi: 10.3389/fopht.2024.1377098. eCollection 2024. Front Ophthalmol (Lausanne). 2024. PMID: 39253560 Free PMC article.
-
Stem Cells, Genome Editing, and the Path to Translational Medicine.Cell. 2018 Oct 18;175(3):615-632. doi: 10.1016/j.cell.2018.09.010. Cell. 2018. PMID: 30340033 Free PMC article. Review.
-
Genome-scale CRISPR screening in a single mouse liver.Cell Genom. 2022 Dec 14;2(12):100217. doi: 10.1016/j.xgen.2022.100217. Epub 2022 Nov 15. Cell Genom. 2022. PMID: 36643909 Free PMC article.
References
-
- Nguyen HT, et al. Gain of 20q11.21 in human embryonic stem cells improves cell survival by increased expression of Bcl-xL. Mol Hum Reprod. 2014;20:168–177. - PubMed
-
- Unger C, Skottman H, Blomberg P, Dilber MS, Hovatta O. Good manufacturing practice and clinical-grade human embryonic stem cell lines. Human Molecular Genetics. 2008;17:R48–53. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

