Optimizing the DNA Donor Template for Homology-Directed Repair of Double-Strand Breaks
- PMID: 28624224
- PMCID: PMC5363683
- DOI: 10.1016/j.omtn.2017.02.006
Optimizing the DNA Donor Template for Homology-Directed Repair of Double-Strand Breaks
Abstract
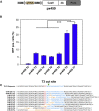
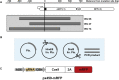
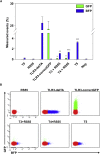
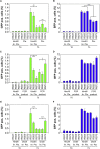
The CRISPR-Cas (clustered regularly interspaced short palindromic repeats-associated proteins) technology enables rapid and precise genome editing at any desired genomic position in almost all cells and organisms. In this study, we analyzed the impact of different repair templates on the frequency of homology-directed repair (HDR) and non-homologous end joining (NHEJ). We used a stable HEK293 cell line expressing the traffic light reporter (TLR-3) system to quantify HDR and NHEJ events following transfection with Cas9, eight different guide RNAs, and a 1,000 bp donor template generated either as circular plasmid, as linearized plasmid with long 3' or 5' backbone overhang, or as PCR product. The sequence to be corrected was either centrally located (RS55), with a shorter 5' homologous region (RS37), or with a shorter 3' homologous region (RS73). Guide RNAs _targeting the transcriptionally active strand (T5, T7) showed significantly higher NHEJ frequencies compared with guide RNAs _targeting the transcriptionally inactive strand. HDR activity was highest when using the linearized plasmid with the short 5' backbone overhang and the RS37 design. The results demonstrate the importance of the design of the guide RNA and template DNA on the frequency of DNA repair events and, ultimately, on the outcome of treatment approaches using HDR.
Keywords: CRISPR-Cas9; DNA donor template; HDR; NHEJ.
Copyright © 2017 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Enhancement of CRISPR-Cas9 induced precise gene editing by _targeting histone H2A-K15 ubiquitination.BMC Biotechnol. 2020 Oct 23;20(1):57. doi: 10.1186/s12896-020-00650-x. BMC Biotechnol. 2020. PMID: 33097066 Free PMC article.
-
Chemical reprogramming enhances homology-directed genome editing in zebrafish embryos.Commun Biol. 2019 May 23;2:198. doi: 10.1038/s42003-019-0444-0. eCollection 2019. Commun Biol. 2019. PMID: 31149642 Free PMC article.
-
Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 with improved proof-reading enhances homology-directed repair.Nucleic Acids Res. 2018 May 18;46(9):4677-4688. doi: 10.1093/nar/gky264. Nucleic Acids Res. 2018. PMID: 29672770 Free PMC article.
-
Advance trends in _targeting homology-directed repair for accurate gene editing: An inclusive review of small molecules and modified CRISPR-Cas9 systems.Bioimpacts. 2022;12(4):371-391. doi: 10.34172/bi.2022.23871. Epub 2022 Jun 22. Bioimpacts. 2022. PMID: 35975201 Free PMC article. Review.
-
Precise Genome Modification via Sequence-Specific Nucleases-Mediated Gene _targeting for Crop Improvement.Front Plant Sci. 2016 Dec 20;7:1928. doi: 10.3389/fpls.2016.01928. eCollection 2016. Front Plant Sci. 2016. PMID: 28066481 Free PMC article. Review.
Cited by
-
An atlas of alternative polyadenylation quantitative trait loci contributing to complex trait and disease heritability.Nat Genet. 2021 Jul;53(7):994-1005. doi: 10.1038/s41588-021-00864-5. Epub 2021 May 13. Nat Genet. 2021. PMID: 33986536
-
3D RNA-scaffolded wireframe origami.Nat Commun. 2023 Jan 24;14(1):382. doi: 10.1038/s41467-023-36156-1. Nat Commun. 2023. PMID: 36693871 Free PMC article.
-
Understanding the diversity of genetic outcomes from CRISPR-Cas generated homology-directed repair.Commun Biol. 2019 Dec 6;2:458. doi: 10.1038/s42003-019-0705-y. eCollection 2019. Commun Biol. 2019. PMID: 31840103 Free PMC article.
-
CRISPR Cpf1 proteins: structure, function and implications for genome editing.Cell Biosci. 2019 May 9;9:36. doi: 10.1186/s13578-019-0298-7. eCollection 2019. Cell Biosci. 2019. PMID: 31086658 Free PMC article. Review.
-
In vitro genome editing rescues parkinsonism phenotypes in induced pluripotent stem cells-derived dopaminergic neurons carrying LRRK2 p.G2019S mutation.Stem Cell Res Ther. 2021 Sep 22;12(1):508. doi: 10.1186/s13287-021-02585-2. Stem Cell Res Ther. 2021. PMID: 34551822 Free PMC article.
References
-
- Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. - PubMed
-
- Barrangou R., van Pijkeren J.P. Exploiting CRISPR-Cas immune systems for genome editing in bacteria. Curr. Opin. Biotechnol. 2016;37:61–68. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

