PAR-TERRA directs homologous sex chromosome pairing
- PMID: 28692038
- PMCID: PMC5553554
- DOI: 10.1038/nsmb.3432
PAR-TERRA directs homologous sex chromosome pairing
Abstract
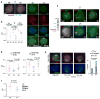
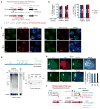
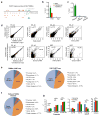
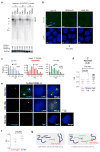
In mammals, homologous chromosomes rarely pair outside meiosis. One exception is the X chromosome, which transiently pairs during X-chromosome inactivation (XCI). How two chromosomes find each other in 3D space is not known. Here, we reveal a required interaction between the X-inactivation center (Xic) and the telomere in mouse embryonic stem (ES) cells. The subtelomeric, pseudoautosomal regions (PARs) of the two sex chromosomes (X and Y) also undergo pairing in both female and male cells. PARs transcribe a class of telomeric RNA, dubbed PAR-TERRA, which accounts for a vast majority of all TERRA transcripts. PAR-TERRA binds throughout the genome, including to the PAR and Xic. During X-chromosome pairing, PAR-TERRA anchors the Xic to the PAR, creating a 'tetrad' of pairwise homologous interactions (Xic-Xic, PAR-PAR, and Xic-PAR). Xic pairing occurs within the tetrad. Depleting PAR-TERRA abrogates pairing and blocks initiation of XCI, whereas autosomal PAR-TERRA induces ectopic pairing. We propose a 'constrained diffusion model' in which PAR-TERRA creates an interaction hub to guide Xic homology searching during XCI.
Figures

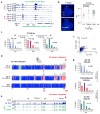
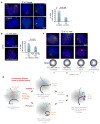
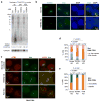
Similar articles
-
Sensing X chromosome pairs before X inactivation via a novel X-pairing region of the Xic.Science. 2007 Dec 7;318(5856):1632-6. doi: 10.1126/science.1149420. Science. 2007. PMID: 18063799
-
PAR-TERRA is the main contributor to telomeric repeat-containing RNA transcripts in normal and cancer mouse cells.RNA. 2021 Jan;27(1):106-121. doi: 10.1261/rna.076281.120. Epub 2020 Oct 30. RNA. 2021. PMID: 33127860 Free PMC article.
-
Ensuring meiotic DNA break formation in the mouse pseudoautosomal region.Nature. 2020 Jun;582(7812):426-431. doi: 10.1038/s41586-020-2327-4. Epub 2020 May 27. Nature. 2020. PMID: 32461690 Free PMC article.
-
The tricky path to recombining X and Y chromosomes in meiosis.Ann N Y Acad Sci. 2012 Sep;1267:18-23. doi: 10.1111/j.1749-6632.2012.06593.x. Ann N Y Acad Sci. 2012. PMID: 22954211 Free PMC article. Review.
-
X inactivation Xplained.Curr Opin Genet Dev. 2007 Oct;17(5):387-93. doi: 10.1016/j.gde.2007.08.001. Epub 2007 Sep 14. Curr Opin Genet Dev. 2007. PMID: 17869504 Review.
Cited by
-
Erdr1 Drives Macrophage Programming via Dynamic Interplay with YAP1 and Mid1.Immunohorizons. 2024 Feb 1;8(2):198-213. doi: 10.4049/immunohorizons.2400004. Immunohorizons. 2024. PMID: 38392560 Free PMC article.
-
XPF activates break-induced telomere synthesis.Nat Commun. 2022 Oct 2;13(1):5781. doi: 10.1038/s41467-022-33428-0. Nat Commun. 2022. PMID: 36184605 Free PMC article.
-
Decapping enzyme 1A breaks X-chromosome symmetry by controlling Tsix elongation and RNA turnover.Nat Cell Biol. 2020 Sep;22(9):1116-1129. doi: 10.1038/s41556-020-0558-0. Epub 2020 Aug 17. Nat Cell Biol. 2020. PMID: 32807903
-
Selective Xi reactivation and alternative methods to restore MECP2 function in Rett syndrome.Trends Genet. 2022 Sep;38(9):920-943. doi: 10.1016/j.tig.2022.01.007. Epub 2022 Mar 2. Trends Genet. 2022. PMID: 35248405 Free PMC article. Review.
-
The Emerging Roles of TERRA in Telomere Maintenance and Genome Stability.Cells. 2019 Mar 15;8(3):246. doi: 10.3390/cells8030246. Cells. 2019. PMID: 30875900 Free PMC article. Review.
References
-
- Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science. 2007;318:798–801. - PubMed
-
- Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nature Cell Biology. 2007;10:228–236. - PubMed
-
- Azzalin CM, Lingner J. Telomere functions grounding on TERRA firma. Trends Cell Biol. 2015;25:29–36. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

