The homeodomain-interacting protein kinase HPK-1 preserves protein homeostasis and longevity through master regulatory control of the HSF-1 chaperone network and TORC1-restricted autophagy in Caenorhabditis elegans
- PMID: 29036198
- PMCID: PMC5658188
- DOI: 10.1371/journal.pgen.1007038
The homeodomain-interacting protein kinase HPK-1 preserves protein homeostasis and longevity through master regulatory control of the HSF-1 chaperone network and TORC1-restricted autophagy in Caenorhabditis elegans
Abstract
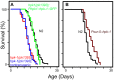
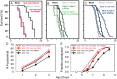
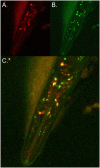
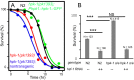
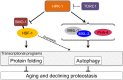
An extensive proteostatic network comprised of molecular chaperones and protein clearance mechanisms functions collectively to preserve the integrity and resiliency of the proteome. The efficacy of this network deteriorates during aging, coinciding with many clinical manifestations, including protein aggregation diseases of the nervous system. A decline in proteostasis can be delayed through the activation of cytoprotective transcriptional responses, which are sensitive to environmental stress and internal metabolic and physiological cues. The homeodomain-interacting protein kinase (hipk) family members are conserved transcriptional co-factors that have been implicated in both genotoxic and metabolic stress responses from yeast to mammals. We demonstrate that constitutive expression of the sole Caenorhabditis elegans Hipk homolog, hpk-1, is sufficient to delay aging, preserve proteostasis, and promote stress resistance, while loss of hpk-1 is deleterious to these phenotypes. We show that HPK-1 preserves proteostasis and extends longevity through distinct but complementary genetic pathways defined by the heat shock transcription factor (HSF-1), and the _target of rapamycin complex 1 (TORC1). We demonstrate that HPK-1 antagonizes sumoylation of HSF-1, a post-translational modification associated with reduced transcriptional activity in mammals. We show that inhibition of sumoylation by RNAi enhances HSF-1-dependent transcriptional induction of chaperones in response to heat shock. We find that hpk-1 is required for HSF-1 to induce molecular chaperones after thermal stress and enhances hormetic extension of longevity. We also show that HPK-1 is required in conjunction with HSF-1 for maintenance of proteostasis in the absence of thermal stress, protecting against the formation of polyglutamine (Q35::YFP) protein aggregates and associated locomotory toxicity. These functions of HPK-1/HSF-1 undergo rapid down-regulation once animals reach reproductive maturity. We show that HPK-1 fortifies proteostasis and extends longevity by an additional independent mechanism: induction of autophagy. HPK-1 is necessary for induction of autophagosome formation and autophagy gene expression in response to dietary restriction (DR) or inactivation of TORC1. The autophagy-stimulating transcription factors pha-4/FoxA and mxl-2/Mlx, but not hlh-30/TFEB or the nuclear hormone receptor nhr-62, are necessary for extended longevity resulting from HPK-1 overexpression. HPK-1 expression is itself induced by transcriptional mechanisms after nutritional stress, and post-transcriptional mechanisms in response to thermal stress. Collectively our results position HPK-1 at a central regulatory node upstream of the greater proteostatic network, acting at the transcriptional level by promoting protein folding via chaperone expression, and protein turnover via expression of autophagy genes. HPK-1 therefore provides a promising intervention point for pharmacological agents _targeting the protein homeostasis system as a means of preserving robust longevity.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Homeodomain-interacting protein kinase maintains neuronal homeostasis during normal Caenorhabditis elegans aging and systemically regulates longevity from serotonergic and GABAergic neurons.Elife. 2023 Jun 20;12:e85792. doi: 10.7554/eLife.85792. Elife. 2023. PMID: 37338980 Free PMC article.
-
TOR Signaling in Caenorhabditis elegans Development, Metabolism, and Aging.Genetics. 2019 Oct;213(2):329-360. doi: 10.1534/genetics.119.302504. Genetics. 2019. PMID: 31594908 Free PMC article. Review.
-
Homeodomain-interacting protein kinase maintains neuronal homeostasis during normal Caenorhabditis elegans aging and systemically regulates longevity from serotonergic and GABAergic neurons.bioRxiv [Preprint]. 2023 Jun 14:2023.01.11.523661. doi: 10.1101/2023.01.11.523661. bioRxiv. 2023. Update in: Elife. 2023 Jun 20;12:e85792. doi: 10.7554/eLife.85792 PMID: 36711523 Free PMC article. Updated. Preprint.
-
Homeodomain-Interacting Protein Kinase (HPK-1) regulates stress responses and ageing in C. elegans.Sci Rep. 2016 Jan 21;6:19582. doi: 10.1038/srep19582. Sci Rep. 2016. PMID: 26791749 Free PMC article.
-
Cellular Homeostasis and Aging.Annu Rev Biochem. 2016 Jun 2;85:1-4. doi: 10.1146/annurev-biochem-011116-110806. Epub 2016 Apr 6. Annu Rev Biochem. 2016. PMID: 27050288 Review.
Cited by
-
Overexpression of homeodomain-interacting protein kinase 2 (HIPK2) attenuates sepsis-mediated liver injury by restoring autophagy.Cell Death Dis. 2018 Aug 28;9(9):847. doi: 10.1038/s41419-018-0838-9. Cell Death Dis. 2018. PMID: 30154452 Free PMC article.
-
HIPK2 Is Required for Midbody Remnant Removal Through Autophagy-Mediated Degradation.Front Cell Dev Biol. 2020 Sep 15;8:572094. doi: 10.3389/fcell.2020.572094. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 33043004 Free PMC article.
-
Homeodomain-interacting protein kinase maintains neuronal homeostasis during normal Caenorhabditis elegans aging and systemically regulates longevity from serotonergic and GABAergic neurons.Elife. 2023 Jun 20;12:e85792. doi: 10.7554/eLife.85792. Elife. 2023. PMID: 37338980 Free PMC article.
-
TOR Signaling in Caenorhabditis elegans Development, Metabolism, and Aging.Genetics. 2019 Oct;213(2):329-360. doi: 10.1534/genetics.119.302504. Genetics. 2019. PMID: 31594908 Free PMC article. Review.
-
Quantifying Tissue-Specific Proteostatic Decline in Caenorhabditis elegans.J Vis Exp. 2021 Sep 7;(175):10.3791/61100. doi: 10.3791/61100. J Vis Exp. 2021. PMID: 34570095 Free PMC article.
References
-
- Kenyon CJ. The genetics of ageing. Nature. 2010;464(7288):504–12. doi: 10.1038/nature08980 . - DOI - PubMed
-
- Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med. 2015;88(Pt B):290–301. doi: 10.1016/j.freeradbiomed.2015.06.008 - DOI - PMC - PubMed
-
- DiLoreto R, Murphy CT. The cell biology of aging. Mol Biol Cell. 2015;26(25):4524–31. doi: 10.1091/mbc.E14-06-1084 - DOI - PMC - PubMed
-
- Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727–54. Epub 2008/04/01. doi: 10.1146/annurev.biochem.77.061206.171059 . - DOI - PubMed
-
- Riera CE, Dillin A. Emerging Role of Sensory Perception in Aging and Metabolism. Trends Endocrinol Metab. 2016;27(5):294–303. doi: 10.1016/j.tem.2016.03.007 . - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

