S-Nitrosylation of PINK1 Attenuates PINK1/Parkin-Dependent Mitophagy in hiPSC-Based Parkinson's Disease Models
- PMID: 29166608
- PMCID: PMC5705204
- DOI: 10.1016/j.celrep.2017.10.068
S-Nitrosylation of PINK1 Attenuates PINK1/Parkin-Dependent Mitophagy in hiPSC-Based Parkinson's Disease Models
Abstract
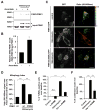
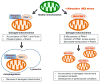
Mutations in PARK6 (PINK1) and PARK2 (Parkin) are linked to rare familial cases of Parkinson's disease (PD). Mutations in these genes result in pathological dysregulation of mitophagy, contributing to neurodegeneration. Here, we report that environmental factors causing a specific posttranslational modification on PINK1 can mimic these genetic mutations. We describe a molecular mechanism for impairment of mitophagy via formation of S-nitrosylated PINK1 (SNO-PINK1). Mitochondrial insults simulating age- or environmental-related stress lead to increased SNO-PINK1, inhibiting its kinase activity. SNO-PINK1 decreases Parkin translocation to mitochondrial membranes, disrupting mitophagy in cell lines and human-iPSC-derived neurons. We find levels of SNO-PINK1 in brains of α-synuclein transgenic PD mice similar to those in cell-based models, indicating the pathophysiological relevance of our findings. Importantly, SNO-PINK1-mediated deficits in mitophagy contribute to neuronal cell death. These results reveal a direct molecular link between nitrosative stress, SNO-PINK1 formation, and mitophagic dysfunction that contributes to the pathogenesis of PD.
Keywords: PARK2; PARK6; PINK1; Parkin; Parkinson’s disease; S-nitrosylation; mitophagy.
Copyright © 2017 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Nix restores mitophagy and mitochondrial function to protect against PINK1/Parkin-related Parkinson's disease.Sci Rep. 2017 Mar 10;7:44373. doi: 10.1038/srep44373. Sci Rep. 2017. PMID: 28281653 Free PMC article.
-
Regulation by mitophagy.Int J Biochem Cell Biol. 2014 Aug;53:147-50. doi: 10.1016/j.biocel.2014.05.012. Epub 2014 May 16. Int J Biochem Cell Biol. 2014. PMID: 24842103 Review.
-
Immunocytochemical Monitoring of PINK1/Parkin-Mediated Mitophagy in Cultured Cells.Methods Mol Biol. 2018;1759:19-27. doi: 10.1007/7651_2017_20. Methods Mol Biol. 2018. PMID: 28361483
-
The PINK1, synphilin-1 and SIAH-1 complex constitutes a novel mitophagy pathway.Hum Mol Genet. 2016 Aug 15;25(16):3476-3490. doi: 10.1093/hmg/ddw189. Epub 2016 Jun 22. Hum Mol Genet. 2016. PMID: 27334109
-
N-degron-mediated degradation and regulation of mitochondrial PINK1 kinase.Curr Genet. 2020 Aug;66(4):693-701. doi: 10.1007/s00294-020-01062-2. Epub 2020 Mar 10. Curr Genet. 2020. PMID: 32157382 Review.
Cited by
-
Oxidative stress and inflammation in osteoarthritis pathogenesis: Role of polyphenols.Biomed Pharmacother. 2020 Sep;129:110452. doi: 10.1016/j.biopha.2020.110452. Epub 2020 Jul 3. Biomed Pharmacother. 2020. PMID: 32768946 Free PMC article. Review.
-
Biochemical study of the effect of mesenchymal stem cells-derived exosome versus L-Dopa in experimentally induced Parkinson's disease in rats.Mol Cell Biochem. 2023 Dec;478(12):2795-2811. doi: 10.1007/s11010-023-04700-8. Epub 2023 Mar 26. Mol Cell Biochem. 2023. PMID: 36966421 Free PMC article.
-
Nutraceuticals _targeting Generation and Oxidant Activity of Peroxynitrite May Aid Prevention and Control of Parkinson's Disease.Int J Mol Sci. 2020 May 21;21(10):3624. doi: 10.3390/ijms21103624. Int J Mol Sci. 2020. PMID: 32455532 Free PMC article. Review.
-
Cell Biology of Parkin: Clues to the Development of New Therapeutics for Parkinson's Disease.CNS Drugs. 2022 Dec;36(12):1249-1267. doi: 10.1007/s40263-022-00973-7. Epub 2022 Nov 15. CNS Drugs. 2022. PMID: 36378485 Review.
-
Organoid and pluripotent stem cells in Parkinson's disease modeling: an expert view on their value to drug discovery.Expert Opin Drug Discov. 2020 Apr;15(4):427-441. doi: 10.1080/17460441.2020.1703671. Epub 2020 Jan 3. Expert Opin Drug Discov. 2020. PMID: 31899983 Free PMC article. Review.
References
-
- Bazan NG. Cell survival matters: docosahexaenoic acid signaling, neuroprotection and photoreceptors. Trends Neurosci. 2006;29:263–271. - PubMed
-
- Bingol B, Tea JS, Phu L, Reichelt M, Bakalarski CE, Song Q, Foreman O, Kirkpatrick DS, Sheng M. The mitochondrial deubiquitinase USP30 opposes Parkin-mediated mitophagy. Nature. 2014;510:370–375. - PubMed
-
- Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, Dawson TM. S-Nitrosylation of parkin regulates ubiquitination and compromises parkin’s protective function. Science. 2004;304:1328–1331. - PubMed
-
- Cociorva D, DLT, Yates JR. Validation of tandem mass spectrometry database search results using DTASelect. Curr Protoc Bioinformatics. 2007;Chapter 13(Unit 13):14. - PubMed
-
- de Vries RL, Przedborski S. Mitophagy and Parkinson’s disease: be eaten to stay healthy. Mol Cell Neurosci. 2013;55:37–43. - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

