Ubiquitination of the Cytoplasmic Domain of Influenza A Virus M2 Protein Is Crucial for Production of Infectious Virus Particles
- PMID: 29167343
- PMCID: PMC5790949
- DOI: 10.1128/JVI.01972-17
Ubiquitination of the Cytoplasmic Domain of Influenza A Virus M2 Protein Is Crucial for Production of Infectious Virus Particles
Abstract
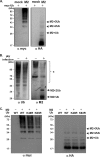
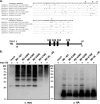
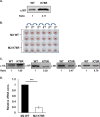
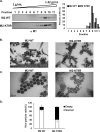
Virus replication is mediated by interactions between the virus and host. Here, we demonstrate that influenza A virus membrane protein 2 (M2) can be ubiquitinated. The lysine residue at position 78, which is located in the cytoplasmic domain of M2, is essential for M2 ubiquitination. An M2-K78R (Lys78→Arg78) mutant, which produces ubiquitination-deficient M2, showed a severe defect in the production of infectious virus particles. M2-K78R mutant progeny contained more hemagglutinin (HA) proteins, less viral RNAs, and less internal viral proteins, including M1 and NP, than the wild-type virus. Furthermore, most of the M2-K78R mutant viral particles lacked viral ribonucleoproteins upon examination by electron microscopy and exhibited slightly lower densities. We also found that mutant M2 colocalized with the M1 protein to a lesser extent than for the wild-type virus. These findings may account for the reduced incorporation of viral ribonucleoprotein into virions. By blocking the second round of virus infection, we showed that the M2 ubiquitination-defective mutant exhibited normal levels of virus replication during the first round of infection, thereby proving that M2 ubiquitination is involved in the virus production step. Finally, we found that the M2-K78R mutant virus induced autophagy and apoptosis earlier than did the wild-type virus. Collectively, these results suggest that M2 ubiquitination plays an important role in infectious virus production by coordinating the efficient packaging of the viral genome into virus particles and the timing of virus-induced cell death.IMPORTANCE Annual epidemics and recurring pandemics of influenza viruses represent very high global health and economic burdens. The influenza virus M2 protein has been extensively studied for its important roles in virus replication, particularly in virus entry and release. Rimantadine, one of the most commonly used antiviral drugs, binds to the channel lumen near the N terminus of M2 proteins. However, viruses that are resistant to rimantadine have emerged. M2 undergoes several posttranslational modifications, such as phosphorylation and palmitoylation. Here, we reveal that ubiquitination mediates the functional role of M2. A ubiquitination-deficient M2 mutant predominately produced virus particles either lacking viral ribonucleoproteins or containing smaller amounts of internal viral components, resulting in lower infectivity. Our findings offer insights into the mechanism of influenza virus morphogenesis, particularly the functional role of M1-M2 interactions in viral particle assembly, and can be applied to the development of new influenza therapies.
Keywords: membrane protein 2; pathogenesis; ubiquitination; virus assembly.
Copyright © 2018 American Society for Microbiology.
Figures
Similar articles
-
Mutations in the Influenza A Virus M1 Protein Enhance Virus Budding To Complement Lethal Mutations in the M2 Cytoplasmic Tail.J Virol. 2017 Dec 14;92(1):e00858-17. doi: 10.1128/JVI.00858-17. Print 2018 Jan 1. J Virol. 2017. PMID: 29046451 Free PMC article.
-
PSMD12-Mediated M1 Ubiquitination of Influenza A Virus at K102 Regulates Viral Replication.J Virol. 2022 Aug 10;96(15):e0078622. doi: 10.1128/jvi.00786-22. Epub 2022 Jul 21. J Virol. 2022. PMID: 35861516 Free PMC article.
-
The influenza A virus M2 cytoplasmic tail is required for infectious virus production and efficient genome packaging.J Virol. 2005 Mar;79(6):3595-605. doi: 10.1128/JVI.79.6.3595-3605.2005. J Virol. 2005. PMID: 15731254 Free PMC article.
-
Influenza virus morphogenesis and budding.Virus Res. 2009 Aug;143(2):147-61. doi: 10.1016/j.virusres.2009.05.010. Epub 2009 May 27. Virus Res. 2009. PMID: 19481124 Free PMC article. Review.
-
Assembly and budding of influenza virus.Virus Res. 2004 Dec;106(2):147-65. doi: 10.1016/j.virusres.2004.08.012. Virus Res. 2004. PMID: 15567494 Free PMC article. Review.
Cited by
-
Flu's cues: Exploiting host post-translational modifications to direct the influenza virus replication cycle.PLoS Pathog. 2018 Sep 20;14(9):e1007205. doi: 10.1371/journal.ppat.1007205. eCollection 2018 Sep. PLoS Pathog. 2018. PMID: 30235357 Free PMC article. Review. No abstract available.
-
Functions of Viroporins in the Viral Life Cycle and Their Regulation of Host Cell Responses.Front Immunol. 2022 Jun 2;13:890549. doi: 10.3389/fimmu.2022.890549. eCollection 2022. Front Immunol. 2022. PMID: 35720341 Free PMC article. Review.
-
Thapsigargin at Non-Cytotoxic Levels Induces a Potent Host Antiviral Response that Blocks Influenza A Virus Replication.Viruses. 2020 Sep 27;12(10):1093. doi: 10.3390/v12101093. Viruses. 2020. PMID: 32992478 Free PMC article.
-
Role of Post-translational Modifications in Influenza A Virus Life Cycle and Host Innate Immune Response.Front Microbiol. 2020 Sep 4;11:517461. doi: 10.3389/fmicb.2020.517461. eCollection 2020. Front Microbiol. 2020. PMID: 33013775 Free PMC article. Review.
-
Protein Palmitoylation Modification During Viral Infection and Detection Methods of Palmitoylated Proteins.Front Cell Infect Microbiol. 2022 Jan 27;12:821596. doi: 10.3389/fcimb.2022.821596. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35155279 Free PMC article. Review.
References
-
- Gannage M, Dormann D, Albrecht R, Dengjel J, Torossi T, Ramer PC, Lee M, Strowig T, Arrey F, Conenello G, Pypaert M, Andersen J, Garcia-Sastre A, Munz C. 2009. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 6:367–380. doi:10.1016/j.chom.2009.09.005. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

