The Deubiquitinase OTU5 Regulates Root Responses to Phosphate Starvation
- PMID: 29301952
- PMCID: PMC5841733
- DOI: 10.1104/pp.17.01525
The Deubiquitinase OTU5 Regulates Root Responses to Phosphate Starvation
Abstract
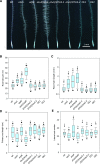
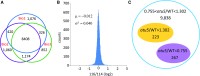
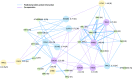
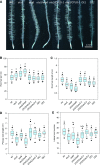
Phosphorus, taken up by plants as inorganic phosphate (Pi), is an essential but often growth-limiting mineral nutrient for plants. As part of an orchestrated response to improve its acquisition, insufficient Pi supply triggers alterations in root architecture and epidermal cell morphogenesis. Arabidopsis (Arabidopsis thaliana) mutants defective in the expression of the OVARIAN TUMOR DOMAIN-CONTAINING DEUBIQUITINATING ENZYME5 (OTU5) exhibited a constitutive Pi deficiency root phenotype, comprising the formation of long and dense root hairs and attenuated primary root growth. Quantitative protein profiling of otu5 and wild-type roots using the isobaric tag for relative and absolute quantification methodology revealed genotype- and Pi-dependent alterations in protein profiles. In otu5 plants, Pi starvation caused a short-root-hair phenotype and decreased abundance of a suite of Pi-responsive root hair-related proteins. Mutant plants also showed the accumulation of proteins involved in chromatin remodeling and altered distribution of reactive oxygen species along the root, which may be causative for the alterations in root hair morphogenesis. The root hair phenotype of otu5 was synergistic to that of actin-related protein6 (arp6), harboring a mutation in the SWR1 chromatin-remodeling complex. Genetic analysis of otu5/arp6 double mutants suggests independent but functionally related roles of the two proteins in chromatin organization. The root hair phenotype of otu5 is not caused by a general up-regulation of the Pi starvation response, indicating that OTU5 acts downstream of or interacts with Pi signaling. It is concluded that OTU5 is involved in the interpretation of environmental information, probably by altering chromatin organization and maintaining redox homeostasis.
© 2018 American Society of Plant Biologists. All Rights Reserved.
Figures

Similar articles
-
Deubiquitinating Enzyme OTU5 Contributes to DNA Methylation Patterns and Is Critical for Phosphate Nutrition Signals.Plant Physiol. 2017 Dec;175(4):1826-1838. doi: 10.1104/pp.17.01188. Epub 2017 Oct 23. Plant Physiol. 2017. PMID: 29061907 Free PMC article.
-
OTU5 tunes environmental responses by sustaining chromatin structure.Plant Signal Behav. 2018 Apr 3;13(4):e1435963. doi: 10.1080/15592324.2018.1435963. Epub 2018 Feb 20. Plant Signal Behav. 2018. PMID: 29393726 Free PMC article.
-
Epigenomic regulation of OTU5 in Arabidopsis thaliana.Genomics. 2020 Sep;112(5):3549-3559. doi: 10.1016/j.ygeno.2020.04.006. Epub 2020 Apr 13. Genomics. 2020. PMID: 32298708
-
Root developmental adaptation to phosphate starvation: better safe than sorry.Trends Plant Sci. 2011 Aug;16(8):442-50. doi: 10.1016/j.tplants.2011.05.006. Trends Plant Sci. 2011. PMID: 21684794 Review.
-
Transcriptional regulation of phosphate acquisition by higher plants.Cell Mol Life Sci. 2012 Oct;69(19):3207-24. doi: 10.1007/s00018-012-1090-6. Epub 2012 Aug 17. Cell Mol Life Sci. 2012. PMID: 22899310 Free PMC article. Review.
Cited by
-
Chromatin enrichment for proteomics in plants (ChEP-P) implicates the histone reader ALFIN-LIKE 6 in jasmonate signalling.BMC Genomics. 2021 Nov 22;22(1):845. doi: 10.1186/s12864-021-08160-6. BMC Genomics. 2021. PMID: 34809577 Free PMC article.
-
Selection and adaptive introgression guided the complex evolutionary history of the European common bean.Nat Commun. 2023 Apr 5;14(1):1908. doi: 10.1038/s41467-023-37332-z. Nat Commun. 2023. PMID: 37019898 Free PMC article.
-
Genetic modification, intercellular communication, and epigenetic regulation in plants: An outlook.Biochem Biophys Res Commun. 2022 Dec 10;633:92-95. doi: 10.1016/j.bbrc.2022.08.044. Epub 2022 Aug 17. Biochem Biophys Res Commun. 2022. PMID: 36344174 Free PMC article. Review. No abstract available.
-
Erasing marks: Functions of plant deubiquitylating enzymes in modulating the ubiquitin code.Plant Cell. 2024 Sep 3;36(9):3057-3073. doi: 10.1093/plcell/koae129. Plant Cell. 2024. PMID: 38656977 Review.
-
Histone Deubiquitinase OTU1 Epigenetically Regulates DA1 and DA2, Which Control Arabidopsis Seed and Organ Size.iScience. 2020 Mar 27;23(3):100948. doi: 10.1016/j.isci.2020.100948. Epub 2020 Feb 28. iScience. 2020. PMID: 32169818 Free PMC article.
References
-
- Berr A, Shafiq S, Pinon V, Dong A, Shen WH (2015) The trxG family histone methyltransferase SET DOMAIN GROUP 26 promotes flowering via a distinctive genetic pathway. Plant J 81: 316–328 - PubMed
-
- Chandrika NN, Sundaravelpandian K, Yu SM, Schmidt W (2013) ALFIN-LIKE 6 is involved in root hair elongation during phosphate deficiency in Arabidopsis. New Phytol 198: 709–720 - PubMed
-
- Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62: 185–206 - PubMed
-
- Choi K, Park C, Lee J, Oh M, Noh B, Lee I (2007) Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134: 1931–1941 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

