WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice
- PMID: 29361035
- PMCID: PMC5861455
- DOI: 10.1093/nar/gky017
WOX11 recruits a histone H3K27me3 demethylase to promote gene expression during shoot development in rice
Abstract
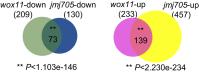
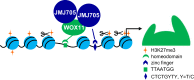
WUSCHEL-related homeobox (WOX) genes are key regulators of meristem activity and plant development, the chromatin mechanism of which to reprogram gene expression remains unclear. Histone H3K27me3 is a chromatin mark of developmentally repressed genes. How the repressive mark is removed from specific genes during plant development is largely unknown. Here, we show that WOX11 interacts with the H3K27me3 demethylase JMJ705 to activate gene expression during shoot development in rice. Genetic analysis indicates that WOX11 and JMJ705 cooperatively control shoot growth and commonly regulate the expression of a set of genes involved in meristem identity, chloroplast biogenesis, and energy metabolism in the shoot apex. Loss of WOX11 led to increased H3K27me3 and overexpression of JMJ705 decreased the methylation levels at a subset of common _targets. JMJ705 is associated with most of the WOX11-binding sites found in the tested common _targets in vivo, regardless of presence or absence of the JMJ705-binding motif. Furthermore, wox11 mutation reduced JMJ705-binding to many _targets genome-wide. The results suggest that recruitment of JMJ705 to specific developmental pathway genes is promoted by DNA-binding transcription factors and that WOX11 functions to stimulate shoot growth through epigenetic reprogramming of genes involved in meristem development and energy-generating pathways.
Figures
Similar articles
-
Rice Homeodomain Protein WOX11 Recruits a Histone Acetyltransferase Complex to Establish Programs of Cell Proliferation of Crown Root Meristem.Plant Cell. 2017 May;29(5):1088-1104. doi: 10.1105/tpc.16.00908. Epub 2017 May 9. Plant Cell. 2017. PMID: 28487409 Free PMC article.
-
The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice.Plant Cell. 2009 Mar;21(3):736-48. doi: 10.1105/tpc.108.061655. Epub 2009 Mar 3. Plant Cell. 2009. PMID: 19258439 Free PMC article.
-
Cooperation between the H3K27me3 Chromatin Mark and Non-CG Methylation in Epigenetic Regulation.Plant Physiol. 2016 Oct;172(2):1131-1141. doi: 10.1104/pp.16.01238. Epub 2016 Aug 17. Plant Physiol. 2016. PMID: 27535791 Free PMC article.
-
Control of shoot cell fate: beyond homeoboxes.Plant Cell. 2001 Apr;13(4):733-8. doi: 10.1105/tpc.13.4.733. Plant Cell. 2001. PMID: 11283332 Free PMC article. Review. No abstract available.
-
Morphogenesis and patterning at the organ boundaries in the higher plant shoot apex.Plant Mol Biol. 2006 Apr;60(6):915-28. doi: 10.1007/s11103-005-2760-7. Plant Mol Biol. 2006. PMID: 16724261 Review.
Cited by
-
Transcription factors WOX11 and LBD16 function with histone demethylase JMJ706 to control crown root development in rice.Plant Cell. 2024 May 1;36(5):1777-1790. doi: 10.1093/plcell/koad318. Plant Cell. 2024. PMID: 38190205 Free PMC article.
-
WUSCHEL-related homeobox genes cooperate with cytokinin to promote bulbil formation in Lilium lancifolium.Plant Physiol. 2022 Aug 29;190(1):387-402. doi: 10.1093/plphys/kiac259. Plant Physiol. 2022. PMID: 35670734 Free PMC article.
-
Ectopic expression of OsWOX9A alters leaf anatomy and plant architecture in rice.Planta. 2024 Jun 16;260(1):30. doi: 10.1007/s00425-024-04463-6. Planta. 2024. PMID: 38879830
-
Genome-wide prediction and functional analysis of WOX genes in blueberry.BMC Genomics. 2024 May 2;25(1):434. doi: 10.1186/s12864-024-10356-5. BMC Genomics. 2024. PMID: 38693497 Free PMC article.
-
Histone Demethylases as Counterbalance to H3K27me3 Silencing in Plants.iScience. 2020 Oct 20;23(11):101715. doi: 10.1016/j.isci.2020.101715. eCollection 2020 Nov 20. iScience. 2020. PMID: 33205025 Free PMC article. Review.
References
-
- Schoof H., Lenhard M., Haecker A., Mayer K.F., Jurgens G., Laux T.. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000; 100:635–644. - PubMed
-
- Leibfried A., To J.P., Busch W., Stehling S., Kehle A., Demar M., Kieber J.J., Lohmann J.U.. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005; 438:1172–1175. - PubMed
-
- Haecker A., Gross-Hardt R., Geiges B., Sarkar A., Breuninger H., Herrmann M., Laux T.. Expression dynamics of WOX genes mark cell fate decisions during early embryonic patterning in Arabidopsis thaliana. Development. 2004; 131:657–668. - PubMed
-
- Wu X.L., Dabi T., Weigel D.. Requirement of homeobox gene STIMPY/WOX9 for Arabidopsis meristem growth and maintenance. Curr. Biol. 2005; 15:436–440. - PubMed
-
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T.. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007; 446:811–814. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

