Development of a Method to Implement Whole-Genome Bisulfite Sequencing of cfDNA from Cancer Patients and a Mouse Tumor Model
- PMID: 29410677
- PMCID: PMC5787102
- DOI: 10.3389/fgene.2018.00006
Development of a Method to Implement Whole-Genome Bisulfite Sequencing of cfDNA from Cancer Patients and a Mouse Tumor Model
Abstract
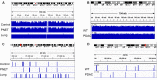
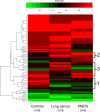
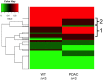
The goal of this study was to develop a method for whole genome cell-free DNA (cfDNA) methylation analysis in humans and mice with the ultimate goal to facilitate the identification of tumor derived DNA methylation changes in the blood. Plasma or serum from patients with pancreatic neuroendocrine tumors or lung cancer, and plasma from a murine model of pancreatic adenocarcinoma was used to develop a protocol for cfDNA isolation, library preparation and whole-genome bisulfite sequencing of ultra low quantities of cfDNA, including tumor-specific DNA. The protocol developed produced high quality libraries consistently generating a conversion rate >98% that will be applicable for the analysis of human and mouse plasma or serum to detect tumor-derived changes in DNA methylation.
Keywords: DNA methylation; biomarker; cell-free DNA; cfDNA; circulating DNA; mouse cfDNA; non-invasive blood based screening; pancreatic cancer.
Figures
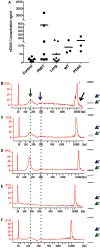
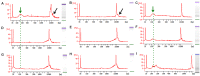
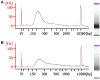
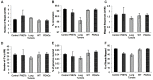
Similar articles
-
Evaluation of commercial kits for isolation and bisulfite conversion of circulating cell-free tumor DNA from blood.Clin Epigenetics. 2023 Sep 14;15(1):151. doi: 10.1186/s13148-023-01563-0. Clin Epigenetics. 2023. PMID: 37710283 Free PMC article.
-
[Practical stability of whole-genome bisulfite sequencing using plasma cell-free DNA].Sheng Wu Gong Cheng Xue Bao. 2019 Dec 25;35(12):2284-2294. doi: 10.13345/j.cjb.190281. Sheng Wu Gong Cheng Xue Bao. 2019. PMID: 31880136 Chinese.
-
Identification of tissue-specific cell death using methylation patterns of circulating DNA.Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):E1826-34. doi: 10.1073/pnas.1519286113. Epub 2016 Mar 14. Proc Natl Acad Sci U S A. 2016. PMID: 26976580 Free PMC article.
-
Using circulating cell-free DNA to monitor personalized cancer therapy.Crit Rev Clin Lab Sci. 2017 May;54(3):205-218. doi: 10.1080/10408363.2017.1299683. Epub 2017 Apr 10. Crit Rev Clin Lab Sci. 2017. PMID: 28393575 Review.
-
Beyond the Blood: CSF-Derived cfDNA for Diagnosis and Characterization of CNS Tumors.Front Cell Dev Biol. 2020 Feb 18;8:45. doi: 10.3389/fcell.2020.00045. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32133357 Free PMC article. Review.
Cited by
-
Low molecular weight serum cell-free DNA concentration is associated with clinicopathologic indices of poor prognosis in women with uterine cancer.J Transl Med. 2020 Aug 27;18(1):323. doi: 10.1186/s12967-020-02493-8. J Transl Med. 2020. PMID: 32854748 Free PMC article.
-
Comparison of commercially available whole-genome sequencing kits for variant detection in circulating cell-free DNA.Sci Rep. 2020 Apr 10;10(1):6190. doi: 10.1038/s41598-020-63102-8. Sci Rep. 2020. PMID: 32277101 Free PMC article.
-
Circulating cell-free methylated DNA reveals tissue-specific, cellular damage from radiation treatment.JCI Insight. 2023 Jul 24;8(14):e156529. doi: 10.1172/jci.insight.156529. JCI Insight. 2023. PMID: 37318863 Free PMC article.
-
Circulating nuclear DNA structural features, origins, and complete size profile revealed by fragmentomics.JCI Insight. 2021 Apr 8;6(7):e144561. doi: 10.1172/jci.insight.144561. JCI Insight. 2021. PMID: 33571170 Free PMC article.
-
Pancreatic Cancer Heterogeneity Can Be Explained Beyond the Genome.Front Oncol. 2019 Apr 5;9:246. doi: 10.3389/fonc.2019.00246. eCollection 2019. Front Oncol. 2019. PMID: 31024848 Free PMC article. Review.
References
-
- Andrews S. R. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

