HLA and proteasome expression body map
- PMID: 29587858
- PMCID: PMC5872580
- DOI: 10.1186/s12920-018-0354-x
HLA and proteasome expression body map
Abstract
Background: The presentation of HLA peptide complexes to T cells is a highly regulated and tissue specific process involving multiple transcriptionally controlled cellular components. The extensive polymorphism of HLA genes and the complex composition of the proteasome make it difficult to map their expression profiles across tissues.
Methods: Here we applied a tailored gene quantification pipeline to 4323 publicly available RNA-Seq datasets representing 55 normal tissues and cell types to examine expression profiles of (classical and non-classical) HLA class I, class II and proteasomal genes.
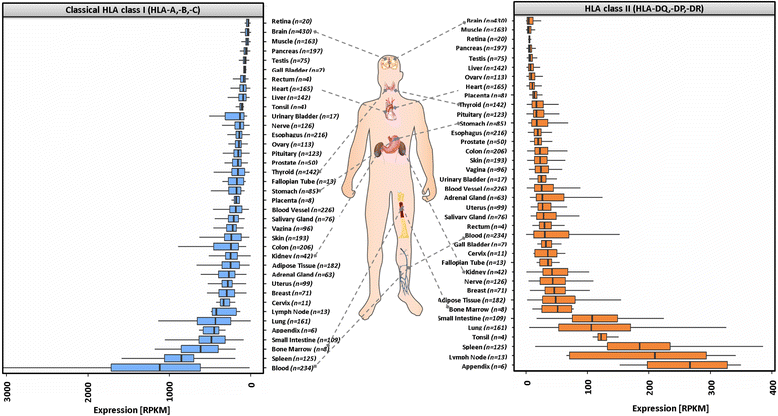
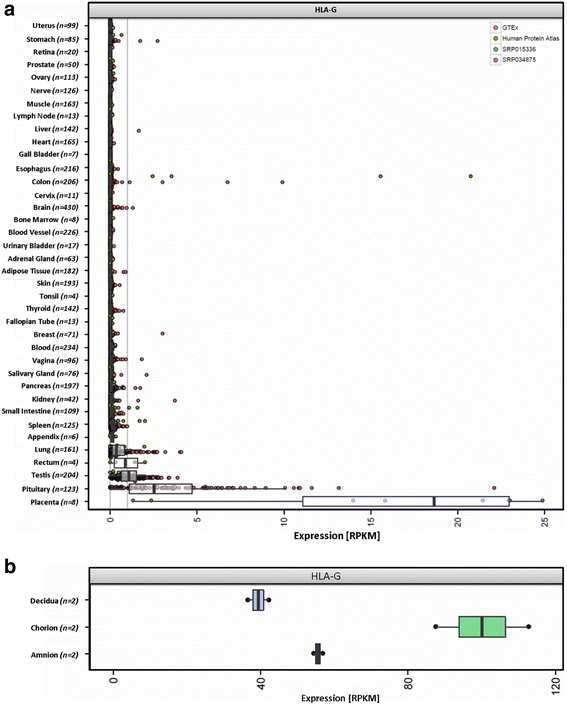
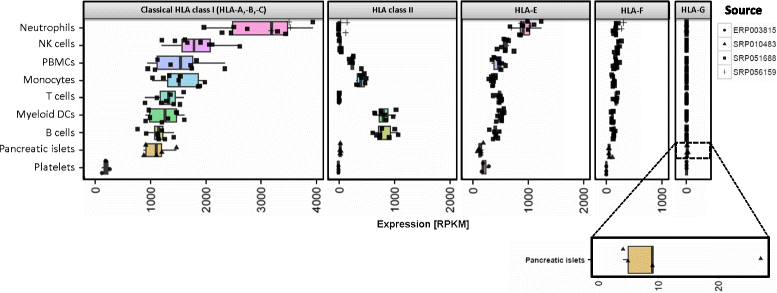
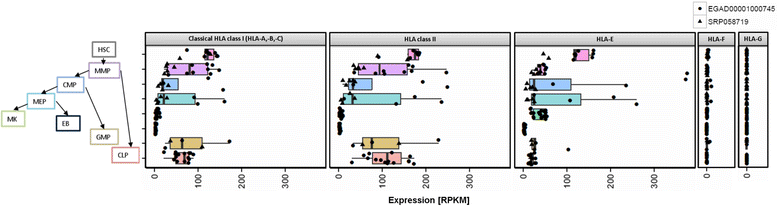
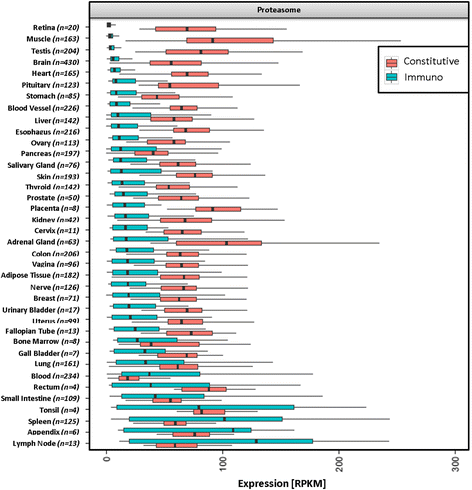
Results: We generated the first comprehensive expression atlas of antigen presenting-related genes across 56 normal tissues and cell types, including immune cells, pancreatic islets, platelets and hematopoietic stem cells. We found a surprisingly heterogeneous HLA expression pattern with up to 100-fold difference in intra-tissue median HLA abundances. Cells of the immune system and lymphatic organs expressed the highest levels of classical HLA class I (HLA-A,-B,-C), class II (HLA-DQA1,-DQB1,-DPA1,-DPB1,-DRA,-DRB1) and non-classical HLA class I (HLA-E,-F) molecules, whereas retina, brain, muscle, megakaryocytes and erythroblasts showed the lowest abundance. In contrast, we identified a distinct and highly tissue-restricted expression pattern of the non-classical class I gene HLA-G in placenta, pancreatic islets, pituitary gland and testis. While the constitutive proteasome showed relatively constant expression across all tissues, we found the immunoproteasome to be enriched in lymphatic organs and almost absent in immune privileged tissues.
Conclusions: Here, we not only provide a reference catalog of tissue and cell type specific HLA expression, but also highlight extremely variable expression of the basic components of antigen processing and presentation in different cell types. Our findings indicate that low expression of classical HLA class I molecules together with lack of immunoproteasome components as well as upregulation of HLA-G may be of key relevance to maintain tolerance in immune privileged tissues.
Keywords: Atlas; Autoimmune; Bioinformatics; HLA expression; Human leukocyte antigens; Immunology; NGS; Proteasome; RNA-Seq.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
US is co-founder and employee of BioNTech AG (Mainz, Germany). JCC is now employed by Agenus UK Limited. These companies are developing immunotherapies. The remaining authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Capturing Differential Allele-Level Expression and Genotypes of All Classical HLA Loci and Haplotypes by a New Capture RNA-Seq Method.Front Immunol. 2020 May 29;11:941. doi: 10.3389/fimmu.2020.00941. eCollection 2020. Front Immunol. 2020. PMID: 32547543 Free PMC article.
-
HLA class I (A, B, C) and class II (DRB1, DQA1, DQB1, DPB1) alleles and haplotypes in the Han from southern China.Tissue Antigens. 2007 Dec;70(6):455-63. doi: 10.1111/j.1399-0039.2007.00932.x. Epub 2007 Sep 27. Tissue Antigens. 2007. PMID: 17900288
-
[Basic understanding of the HLA system in allogeneic hematopoietic cell transplantation].Rinsho Ketsueki. 2015 Oct;56(10):2134-43. doi: 10.11406/rinketsu.56.2134. Rinsho Ketsueki. 2015. PMID: 26458453 Japanese.
-
The HLA genomic loci map: expression, interaction, diversity and disease.J Hum Genet. 2009 Jan;54(1):15-39. doi: 10.1038/jhg.2008.5. Epub 2009 Jan 9. J Hum Genet. 2009. PMID: 19158813 Review.
-
[Non-classical human leukocyte antigen (HLA) tissue types--from implantation to transplantation].Ugeskr Laeger. 2006 Jan 30;168(5):461-6. Ugeskr Laeger. 2006. PMID: 16472433 Review. Danish.
Cited by
-
HLA allele-specific expression: Methods, disease associations, and relevance in hematopoietic stem cell transplantation.Front Immunol. 2022 Sep 28;13:1007425. doi: 10.3389/fimmu.2022.1007425. eCollection 2022. Front Immunol. 2022. PMID: 36248878 Free PMC article. Review.
-
Deciphering the HLA-E immunopeptidome with mass spectrometry: an opportunity for universal mRNA vaccines and T-cell-directed immunotherapies.Front Immunol. 2024 Sep 5;15:1442783. doi: 10.3389/fimmu.2024.1442783. eCollection 2024. Front Immunol. 2024. PMID: 39301027 Free PMC article. Review.
-
An Insight into Recent Advances on Platelet Function in Health and Disease.Int J Mol Sci. 2022 May 27;23(11):6022. doi: 10.3390/ijms23116022. Int J Mol Sci. 2022. PMID: 35682700 Free PMC article. Review.
-
CRISPR activation enables high-fidelity reprogramming into human pluripotent stem cells.Stem Cell Reports. 2022 Feb 8;17(2):413-426. doi: 10.1016/j.stemcr.2021.12.017. Epub 2022 Jan 20. Stem Cell Reports. 2022. PMID: 35063129 Free PMC article.
-
Polarized HLA Class I Expression on Renal Tubules Hinders the Detection of Donor-Specific Urinary Extracellular Vesicles.Int J Nanomedicine. 2024 Apr 12;19:3497-3511. doi: 10.2147/IJN.S446525. eCollection 2024. Int J Nanomedicine. 2024. PMID: 38628433 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

