Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer
- PMID: 29615789
- PMCID: PMC5895503
- DOI: 10.1038/s41586-018-0018-1
Metabolic enzyme PFKFB4 activates transcriptional coactivator SRC-3 to drive breast cancer
Abstract
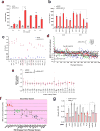
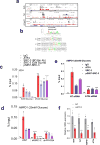
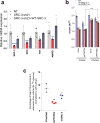
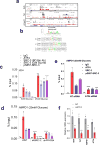
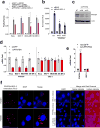
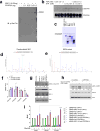
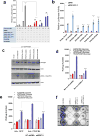
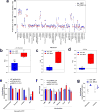
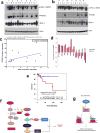
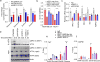
Alterations in both cell metabolism and transcriptional programs are hallmarks of cancer that sustain rapid proliferation and metastasis 1 . However, the mechanisms that control the interaction between metabolic reprogramming and transcriptional regulation remain unclear. Here we show that the metabolic enzyme 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4) regulates transcriptional reprogramming by activating the oncogenic steroid receptor coactivator-3 (SRC-3). We used a kinome-wide RNA interference-based screening method to identify potential kinases that modulate the intrinsic SRC-3 transcriptional response. PFKFB4, a regulatory enzyme that synthesizes a potent stimulator of glycolysis 2 , is found to be a robust stimulator of SRC-3 that coregulates oestrogen receptor. PFKFB4 phosphorylates SRC-3 at serine 857 and enhances its transcriptional activity, whereas either suppression of PFKFB4 or ectopic expression of a phosphorylation-deficient Ser857Ala mutant SRC-3 abolishes the SRC-3-mediated transcriptional output. Functionally, PFKFB4-driven SRC-3 activation drives glucose flux towards the pentose phosphate pathway and enables purine synthesis by transcriptionally upregulating the expression of the enzyme transketolase. In addition, the two enzymes adenosine monophosphate deaminase-1 (AMPD1) and xanthine dehydrogenase (XDH), which are involved in purine metabolism, were identified as SRC-3 _targets that may or may not be directly involved in purine synthesis. Mechanistically, phosphorylation of SRC-3 at Ser857 increases its interaction with the transcription factor ATF4 by stabilizing the recruitment of SRC-3 and ATF4 to _target gene promoters. Ablation of SRC-3 or PFKFB4 suppresses breast tumour growth in mice and prevents metastasis to the lung from an orthotopic setting, as does Ser857Ala-mutant SRC-3. PFKFB4 and phosphorylated SRC-3 levels are increased and correlate in oestrogen receptor-positive tumours, whereas, in patients with the basal subtype, PFKFB4 and SRC-3 drive a common protein signature that correlates with the poor survival of patients with breast cancer. These findings suggest that the Warburg pathway enzyme PFKFB4 acts as a molecular fulcrum that couples sugar metabolism to transcriptional activation by stimulating SRC-3 to promote aggressive metastatic tumours.
Figures




Comment in
-
A Glycolysis Outsider Steps into the Cancer Spotlight.Cell Metab. 2018 Jul 3;28(1):3-4. doi: 10.1016/j.cmet.2018.06.017. Cell Metab. 2018. PMID: 29972796
Similar articles
-
PFKFB4 promotes lung adenocarcinoma progression via phosphorylating and activating transcriptional coactivator SRC-2.BMC Pulm Med. 2021 Feb 16;21(1):60. doi: 10.1186/s12890-021-01420-x. BMC Pulm Med. 2021. PMID: 33593309 Free PMC article.
-
6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 is essential for p53-null cancer cells.Oncogene. 2017 Jun 8;36(23):3287-3299. doi: 10.1038/onc.2016.477. Epub 2017 Jan 16. Oncogene. 2017. PMID: 28092678
-
6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 and 4: A pair of valves for fine-tuning of glucose metabolism in human cancer.Mol Metab. 2019 Feb;20:1-13. doi: 10.1016/j.molmet.2018.11.013. Epub 2018 Dec 5. Mol Metab. 2019. PMID: 30553771 Free PMC article. Review.
-
PELP1/SRC-3-dependent regulation of metabolic PFKFB kinases drives therapy resistant ER+ breast cancer.Oncogene. 2021 Jun;40(25):4384-4397. doi: 10.1038/s41388-021-01871-w. Epub 2021 Jun 8. Oncogene. 2021. PMID: 34103681 Free PMC article.
-
Nuances of PFKFB3 Signaling in Breast Cancer.Clin Breast Cancer. 2022 Jun;22(4):e604-e614. doi: 10.1016/j.clbc.2022.01.002. Epub 2022 Jan 15. Clin Breast Cancer. 2022. PMID: 35135735 Review.
Cited by
-
SRC-3, a Steroid Receptor Coactivator: Implication in Cancer.Int J Mol Sci. 2021 Apr 30;22(9):4760. doi: 10.3390/ijms22094760. Int J Mol Sci. 2021. PMID: 33946224 Free PMC article. Review.
-
Traditional Herbal Medicine: A Potential Therapeutic Approach for Adjuvant Treatment of Non-small Cell Lung Cancer in the Future.Integr Cancer Ther. 2022 Jan-Dec;21:15347354221144312. doi: 10.1177/15347354221144312. Integr Cancer Ther. 2022. PMID: 36567455 Free PMC article. Review.
-
The enhancement of glycolysis regulates pancreatic cancer metastasis.Cell Mol Life Sci. 2020 Jan;77(2):305-321. doi: 10.1007/s00018-019-03278-z. Epub 2019 Aug 20. Cell Mol Life Sci. 2020. PMID: 31432232 Free PMC article. Review.
-
Current Status of the Use of Multifunctional Enzymes as Anti-Cancer Drug _targets.Pharmaceutics. 2021 Dec 21;14(1):10. doi: 10.3390/pharmaceutics14010010. Pharmaceutics. 2021. PMID: 35056904 Free PMC article. Review.
-
THOC3 interacts with YBX1 to promote lung squamous cell carcinoma progression through PFKFB4 mRNA modification.Cell Death Dis. 2023 Jul 27;14(7):475. doi: 10.1038/s41419-023-06008-3. Cell Death Dis. 2023. PMID: 37500615 Free PMC article.
References
-
- Dang CV. Cancer cell metabolism: there is no ROS for the weary. Cancer. Discov. 2012;2:304–307. - PubMed
-
- Gojis O, et al. The role of SRC-3 in human breast cancer. Nat. Rev. Clin. Oncol. 2010;7:83–89. - PubMed
-
- Anzick SL, et al. AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science. 1997;277:965–968. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

