Regulation of chromatin and gene expression by metabolic enzymes and metabolites
- PMID: 29930302
- PMCID: PMC6907087
- DOI: 10.1038/s41580-018-0029-7
Regulation of chromatin and gene expression by metabolic enzymes and metabolites
Abstract
Metabolism and gene expression, which are two fundamental biological processes that are essential to all living organisms, reciprocally regulate each other to maintain homeostasis and regulate cell growth, survival and differentiation. Metabolism feeds into the regulation of gene expression via metabolic enzymes and metabolites, which can modulate chromatin directly or indirectly - through regulation of the activity of chromatin trans-acting proteins, including histone-modifying enzymes, chromatin-remodelling complexes and transcription regulators. Deregulation of these metabolic activities has been implicated in human diseases, prominently including cancer.
Conflict of interest statement
Competing interests
The authors declare no competing interests.
Figures

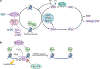

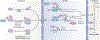
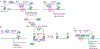
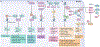
Similar articles
-
Rheostat control of gene expression by metabolites.Mol Cell. 2006 Oct 6;24(1):1-11. doi: 10.1016/j.molcel.2006.09.002. Mol Cell. 2006. PMID: 17018288 Review.
-
Modifying Chromatin by Histone Tail Clipping.J Mol Biol. 2018 Sep 14;430(18 Pt B):3051-3067. doi: 10.1016/j.jmb.2018.07.013. Epub 2018 Jul 25. J Mol Biol. 2018. PMID: 30009770 Review.
-
Enhancer function regulated by combinations of transcription factors and cofactors.Genes Cells. 2018 Oct;23(10):808-821. doi: 10.1111/gtc.12634. Epub 2018 Aug 31. Genes Cells. 2018. PMID: 30092612 Review.
-
Experimental analysis of chromatin function in transcription control.Crit Rev Eukaryot Gene Expr. 1994;4(4):403-41. Crit Rev Eukaryot Gene Expr. 1994. PMID: 7734837 Review.
-
Interplay between chromatin remodelers and protein arginine methyltransferases.J Cell Physiol. 2007 Nov;213(2):306-15. doi: 10.1002/jcp.21180. J Cell Physiol. 2007. PMID: 17708529 Review.
Cited by
-
The evolving metabolic landscape of chromatin biology and epigenetics.Nat Rev Genet. 2020 Dec;21(12):737-753. doi: 10.1038/s41576-020-0270-8. Epub 2020 Sep 9. Nat Rev Genet. 2020. PMID: 32908249 Free PMC article. Review.
-
Oxidative stress-initiated one-carbon metabolism drives the generation of interleukin-10-producing B cells to resolve pneumonia.Cell Mol Immunol. 2024 Jan;21(1):19-32. doi: 10.1038/s41423-023-01109-7. Epub 2023 Dec 11. Cell Mol Immunol. 2024. PMID: 38082147 Free PMC article.
-
Acylations in cardiovascular biology and diseases, what's beyond acetylation.EBioMedicine. 2023 Jan;87:104418. doi: 10.1016/j.ebiom.2022.104418. Epub 2022 Dec 28. EBioMedicine. 2023. PMID: 36584593 Free PMC article. Review.
-
HIF3A Inhibition Triggers Browning of White Adipocytes via Metabolic Rewiring.Front Cell Dev Biol. 2022 Jan 12;9:740203. doi: 10.3389/fcell.2021.740203. eCollection 2021. Front Cell Dev Biol. 2022. PMID: 35096807 Free PMC article.
-
A metabolic perspective of the neutrophil life cycle: new avenues in immunometabolism.Front Immunol. 2024 Jan 8;14:1334205. doi: 10.3389/fimmu.2023.1334205. eCollection 2023. Front Immunol. 2024. PMID: 38259490 Free PMC article. Review.
References
-
- Luger K, Mader AW, Richmond RK, Sargent DF & Richmond TJ Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997). - PubMed
-
- Strahl BD & Allis CD The language of covalent histone modifications. Nature 403, 41–45 (2000). - PubMed
-
-
Tan M et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 146, 1016–1028 (2011).
This study describes a large number of novel histone marks, including lysine crotonylation and tyrosine hydroxylation as novel histone modifications.
-
-
- Tessarz P & Kouzarides T Histone core modifications regulating nucleosome structure and dynamics. Nat. Rev. Mol. Cell Biol 15, 703–708 (2014). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

