Consensus molecular subtype classification of colorectal adenomas
- PMID: 29968252
- PMCID: PMC6221003
- DOI: 10.1002/path.5129
Consensus molecular subtype classification of colorectal adenomas
Abstract
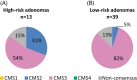
Consensus molecular subtyping is an RNA expression-based classification system for colorectal cancer (CRC). Genomic alterations accumulate during CRC pathogenesis, including the premalignant adenoma stage, leading to changes in RNA expression. Only a minority of adenomas progress to malignancies, a transition that is associated with specific DNA copy number aberrations or microsatellite instability (MSI). We aimed to investigate whether colorectal adenomas can already be stratified into consensus molecular subtype (CMS) classes, and whether specific CMS classes are related to the presence of specific DNA copy number aberrations associated with progression to malignancy. RNA sequencing was performed on 62 adenomas and 59 CRCs. MSI status was determined with polymerase chain reaction-based methodology. DNA copy number was assessed by low-coverage DNA sequencing (n = 30) or array-comparative genomic hybridisation (n = 32). Adenomas were classified into CMS classes together with CRCs from the study cohort and from The Cancer Genome Atlas (n = 556), by use of the established CMS classifier. As a result, 54 of 62 (87%) adenomas were classified according to the CMS. The CMS3 'metabolic subtype', which was least common among CRCs, was most prevalent among adenomas (n = 45; 73%). One of the two adenomas showing MSI was classified as CMS1 (2%), the 'MSI immune' subtype. Eight adenomas (13%) were classified as the 'canonical' CMS2. No adenomas were classified as the 'mesenchymal' CMS4, consistent with the fact that adenomas lack invasion-associated stroma. The distribution of the CMS classes among adenomas was confirmed in an independent series. CMS3 was enriched with adenomas at low risk of progressing to CRC, whereas relatively more high-risk adenomas were observed in CMS2. We conclude that adenomas can be stratified into the CMS classes. Considering that CMS1 and CMS2 expression signatures may mark adenomas at increased risk of progression, the distribution of the CMS classes among adenomas is consistent with the proportion of adenomas expected to progress to CRC. © 2018 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Keywords: adenoma; colon; colorectal cancer; neoplasia; rectum.
© 2018 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Figures

Similar articles
-
Molecular characterization of colorectal adenomas reveals POFUT1 as a candidate driver of tumor progression.Int J Cancer. 2020 Apr 1;146(7):1979-1992. doi: 10.1002/ijc.32627. Epub 2019 Aug 30. Int J Cancer. 2020. PMID: 31411736 Free PMC article.
-
Colorectal premalignancy is associated with consensus molecular subtypes 1 and 2.Ann Oncol. 2018 Oct 1;29(10):2061-2067. doi: 10.1093/annonc/mdy337. Ann Oncol. 2018. PMID: 30412224 Free PMC article.
-
Consensus molecular subtype transition during progression of colorectal cancer.J Pathol. 2023 Nov;261(3):298-308. doi: 10.1002/path.6176. Epub 2023 Sep 8. J Pathol. 2023. PMID: 37681286
-
Molecular pathological classification of colorectal cancer.Virchows Arch. 2016 Aug;469(2):125-34. doi: 10.1007/s00428-016-1956-3. Epub 2016 Jun 20. Virchows Arch. 2016. PMID: 27325016 Free PMC article. Review.
-
Back to the Colorectal Cancer Consensus Molecular Subtype Future.Curr Gastroenterol Rep. 2019 Jan 30;21(2):5. doi: 10.1007/s11894-019-0674-9. Curr Gastroenterol Rep. 2019. PMID: 30701321 Free PMC article. Review.
Cited by
-
Classification of tumor types using XGBoost machine learning model: a vector space transformation of genomic alterations.J Transl Med. 2023 Nov 21;21(1):836. doi: 10.1186/s12967-023-04720-4. J Transl Med. 2023. PMID: 37990214 Free PMC article.
-
Suspected Hereditary Cancer Syndromes in Young Patients: Heterogeneous Clinical and Genetic Presentation of Colorectal Cancers.Oncologist. 2019 Jul;24(7):877-882. doi: 10.1634/theoncologist.2018-0614. Epub 2019 Jan 25. Oncologist. 2019. PMID: 30683709 Free PMC article.
-
Principles of Molecular Utility for CMS Classification in Colorectal Cancer Management.Cancers (Basel). 2023 May 13;15(10):2746. doi: 10.3390/cancers15102746. Cancers (Basel). 2023. PMID: 37345083 Free PMC article. Review.
-
Identification of modules and functional analysis in CRC subtypes by integrated bioinformatics analysis.PLoS One. 2019 Aug 30;14(8):e0221772. doi: 10.1371/journal.pone.0221772. eCollection 2019. PLoS One. 2019. PMID: 31469863 Free PMC article.
-
Association Between Tumor Mutation Profile and Clinical Outcomes Among Hispanic-Latino Patients With Metastatic Colorectal Cancer.Front Oncol. 2022 Jan 24;11:772225. doi: 10.3389/fonc.2021.772225. eCollection 2021. Front Oncol. 2022. PMID: 35141142 Free PMC article.
References
-
- De Sousa EMF, Wang X, Jansen M, et al Poor‐prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med 2013; 19 : 614–618. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

