Plasma proteomic signature of age in healthy humans
- PMID: 29992704
- PMCID: PMC6156492
- DOI: 10.1111/acel.12799
Plasma proteomic signature of age in healthy humans
Abstract
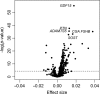
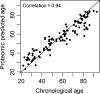
To characterize the proteomic signature of chronological age, 1,301 proteins were measured in plasma using the SOMAscan assay (SomaLogic, Boulder, CO, USA) in a population of 240 healthy men and women, 22-93 years old, who were disease- and treatment-free and had no physical and cognitive impairment. Using a p ≤ 3.83 × 10-5 significance threshold, 197 proteins were positively associated, and 20 proteins were negatively associated with age. Growth differentiation factor 15 (GDF15) had the strongest, positive association with age (GDF15; 0.018 ± 0.001, p = 7.49 × 10-56 ). In our sample, GDF15 was not associated with other cardiovascular risk factors such as cholesterol or inflammatory markers. The functional pathways enriched in the 217 age-associated proteins included blood coagulation, chemokine and inflammatory pathways, axon guidance, peptidase activity, and apoptosis. Using elastic net regression models, we created a proteomic signature of age based on relative concentrations of 76 proteins that highly correlated with chronological age (r = 0.94). The generalizability of our findings needs replication in an independent cohort.
Keywords: aging; aptamers; healthy aging; humans; plasma; proteomics.
© 2018 The Authors. Aging Cell published by the Anatomical Society and John Wiley & Sons Ltd.
Figures


Similar articles
-
Plasma Proteomic Signature of Cellular Senescence and Markers of Biological Aging Among Postmenopausal Women.Rejuvenation Res. 2022 Jun;25(3):141-148. doi: 10.1089/rej.2022.0024. Epub 2022 Jun 3. Rejuvenation Res. 2022. PMID: 35583231
-
Plasma proteomic signature of human longevity.Aging Cell. 2024 Jun;23(6):e14136. doi: 10.1111/acel.14136. Epub 2024 Mar 5. Aging Cell. 2024. PMID: 38440820 Free PMC article.
-
Circulating Proteomic Signatures of Chronological Age.J Gerontol A Biol Sci Med Sci. 2015 Jul;70(7):809-16. doi: 10.1093/gerona/glu121. Epub 2014 Aug 14. J Gerontol A Biol Sci Med Sci. 2015. PMID: 25123647 Free PMC article.
-
Habitual aerobic exercise and circulating proteomic patterns in healthy adults: relation to indicators of healthspan.J Appl Physiol (1985). 2018 Nov 1;125(5):1646-1659. doi: 10.1152/japplphysiol.00458.2018. Epub 2018 Sep 20. J Appl Physiol (1985). 2018. PMID: 30236049 Free PMC article.
-
Proteomics in aging research: A roadmap to clinical, translational research.Aging Cell. 2021 Apr;20(4):e13325. doi: 10.1111/acel.13325. Epub 2021 Mar 17. Aging Cell. 2021. PMID: 33730416 Free PMC article. Review.
Cited by
-
Plasma GDF-15 concentration is not elevated in open-angle glaucoma.PLoS One. 2021 May 28;16(5):e0252630. doi: 10.1371/journal.pone.0252630. eCollection 2021. PLoS One. 2021. PMID: 34048486 Free PMC article.
-
Podocyte senescence: from molecular mechanisms to therapeutics.Ren Fail. 2024 Dec;46(2):2398712. doi: 10.1080/0886022X.2024.2398712. Epub 2024 Sep 9. Ren Fail. 2024. PMID: 39248407 Free PMC article. Review.
-
The social nature of mitochondria: Implications for human health.Neurosci Biobehav Rev. 2021 Jan;120:595-610. doi: 10.1016/j.neubiorev.2020.04.017. Epub 2020 Jul 8. Neurosci Biobehav Rev. 2021. PMID: 32651001 Free PMC article. Review.
-
Minimum standards for evaluating machine-learned models of high-dimensional data.Front Aging. 2022 Sep 13;3:901841. doi: 10.3389/fragi.2022.901841. eCollection 2022. Front Aging. 2022. PMID: 36176975 Free PMC article.
-
Influence of Growth Differentiation Factor 15 on Intraocular Pressure in Mice.Lab Invest. 2024 Apr;104(4):102025. doi: 10.1016/j.labinv.2024.102025. Epub 2024 Jan 28. Lab Invest. 2024. PMID: 38290601
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

