Phosphorylation of ULK1 affects autophagosome fusion and links chaperone-mediated autophagy to macroautophagy
- PMID: 30154410
- PMCID: PMC6113293
- DOI: 10.1038/s41467-018-05449-1
Phosphorylation of ULK1 affects autophagosome fusion and links chaperone-mediated autophagy to macroautophagy
Abstract
The Unc-51 like autophagy activating kinase 1 (ULK1) complex plays a central role in the initiation stage of autophagy. However, the function of ULK1 in the late stage of autophagy is unknown. Here, we report that ULK1, a central kinase of the ULK1 complex involved in autophagy initiation, promotes autophagosome-lysosome fusion. PKCα phosphorylates ULK1 and prevents autolysosome formation. PKCα phosphorylation of ULK1 does not change its kinase activity; however, it decreases autophagosome-lysosome fusion by reducing the affinity of ULK1 for syntaxin 17 (STX17). Unphosphorylated ULK1 recruited STX17 and increased STX17's affinity towards synaptosomal-associated protein 29 (SNAP29). Additionally, phosphorylation of ULK1 enhances its interaction with heat shock cognate 70 kDa protein (HSC70) and increases its degradation through chaperone-mediated autophagy (CMA). Our study unearths a key mechanism underlying autolysosome formation, a process in which the kinase activity of PKCα plays an instrumental role, and reveals the significance of the mutual regulation of macroautophagy and CMA in maintaining the balance of autophagy.
Conflict of interest statement
The authors declare no competing interests.
Figures


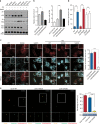
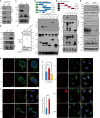
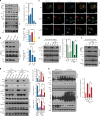
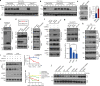

Similar articles
-
The Vici Syndrome Protein EPG5 Is a Rab7 Effector that Determines the Fusion Specificity of Autophagosomes with Late Endosomes/Lysosomes.Mol Cell. 2016 Sep 1;63(5):781-95. doi: 10.1016/j.molcel.2016.08.021. Mol Cell. 2016. PMID: 27588602
-
RUNDC1 inhibits autolysosome formation and survival of zebrafish via clasping ATG14-STX17-SNAP29 complex.Cell Death Differ. 2023 Oct;30(10):2231-2248. doi: 10.1038/s41418-023-01215-z. Epub 2023 Sep 8. Cell Death Differ. 2023. PMID: 37684417 Free PMC article.
-
New insights regarding SNARE proteins in autophagosome-lysosome fusion.Autophagy. 2021 Oct;17(10):2680-2688. doi: 10.1080/15548627.2020.1823124. Epub 2020 Sep 24. Autophagy. 2021. PMID: 32924745 Free PMC article. Review.
-
Aberrant autophagosome formation occurs upon small molecule inhibition of ULK1 kinase activity.Life Sci Alliance. 2020 Oct 27;3(12):e202000815. doi: 10.26508/lsa.202000815. Print 2020 Dec. Life Sci Alliance. 2020. PMID: 33109685 Free PMC article.
-
Regulation of Syntaxin 17 during Autophagosome Maturation.Trends Cell Biol. 2019 Jan;29(1):1-3. doi: 10.1016/j.tcb.2018.10.003. Epub 2018 Nov 8. Trends Cell Biol. 2019. PMID: 30415939 Review.
Cited by
-
Integration of O-GlcNAc into Stress Response Pathways.Cells. 2022 Nov 5;11(21):3509. doi: 10.3390/cells11213509. Cells. 2022. PMID: 36359905 Free PMC article. Review.
-
A Review of ULK1-Mediated Autophagy in Drug Resistance of Cancer.Cancers (Basel). 2020 Feb 4;12(2):352. doi: 10.3390/cancers12020352. Cancers (Basel). 2020. PMID: 32033142 Free PMC article. Review.
-
Protein and Mitochondria Quality Control Mechanisms and Cardiac Aging.Cells. 2020 Apr 10;9(4):933. doi: 10.3390/cells9040933. Cells. 2020. PMID: 32290135 Free PMC article. Review.
-
The Long Noncoding RNA LOC441461 (STX17-AS1) Modulates Colorectal Cancer Cell Growth and Motility.Cancers (Basel). 2020 Oct 28;12(11):3171. doi: 10.3390/cancers12113171. Cancers (Basel). 2020. PMID: 33126743 Free PMC article.
-
β-Arrestin2 Is Critically Involved in the Differential Regulation of Phosphosignaling Pathways by Thyrotropin-Releasing Hormone and Taltirelin.Cells. 2022 Apr 27;11(9):1473. doi: 10.3390/cells11091473. Cells. 2022. PMID: 35563779 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

