Dynamics and functional interplay of histone lysine butyrylation, crotonylation, and acetylation in rice under starvation and submergence
- PMID: 30253806
- PMCID: PMC6154804
- DOI: 10.1186/s13059-018-1533-y
Dynamics and functional interplay of histone lysine butyrylation, crotonylation, and acetylation in rice under starvation and submergence
Abstract
Background: Histone lysine acylations by short-chain fatty acids are distinct from the widely studied histone lysine acetylation in chromatin, although both modifications are regulated by primary metabolism in mammalian cells. It remains unknown whether and how histone acylation and acetylation interact to regulate gene expression in plants that have distinct regulatory pathways of primary metabolism.
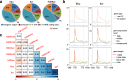
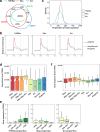
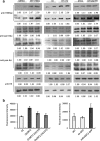
Results: We identify 4 lysine butyrylation (Kbu) sites (H3K14, H4K12, H2BK42, and H2BK134) and 45 crotonylation (Kcr) sites on rice histones by mass spectrometry. Comparative analysis of genome-wide Kbu and Kcr and H3K9ac in combination with RNA sequencing reveals 25,306 genes marked by Kbu and Kcr in rice and more than 95% of H3K9ac-marked genes are marked by both. Kbu and Kcr are enriched at the 5' region of expressed genes. In rice under starvation and submergence, Kbu and Kcr appear to be less dynamic and display changes in different sets of genes compared to H3K9ac. Furthermore, Kbu seems to preferentially poise gene activation by external stresses, rather than internal circadian rhythm which has been shown to be tightly associated with H3K9ac. In addition, we show that rice sirtuin histone deacetylase (SRT2) is involved in the removal of Kcr.
Conclusion: Kbu, Kcr, and H3K9ac redundantly mark a large number of active genes but display different responses to external and internal signals. Thus, the proportion of rice histone lysine acetylation and acylation is dynamically regulated by environmental and metabolic cues, which may represent an epigenetic mechanism to fine-tune gene expression for plant adaptation.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



Similar articles
-
Global Involvement of Lysine Crotonylation in Protein Modification and Transcription Regulation in Rice.Mol Cell Proteomics. 2018 Oct;17(10):1922-1936. doi: 10.1074/mcp.RA118.000640. Epub 2018 Jul 18. Mol Cell Proteomics. 2018. PMID: 30021883 Free PMC article.
-
Genome-wide Profiling of Histone Lysine Butyrylation Reveals its Role in the Positive Regulation of Gene Transcription in Rice.Rice (N Y). 2019 Nov 27;12(1):86. doi: 10.1186/s12284-019-0342-6. Rice (N Y). 2019. PMID: 31776817 Free PMC article.
-
Dynamic profiling and functional interpretation of histone lysine crotonylation and lactylation during neural development.Development. 2022 Jul 15;149(14):dev200049. doi: 10.1242/dev.200049. Epub 2022 Jul 21. Development. 2022. PMID: 35735108
-
Lysine crotonylation: A challenging new player in the epigenetic regulation of plants.J Proteomics. 2022 Mar 20;255:104488. doi: 10.1016/j.jprot.2022.104488. Epub 2022 Jan 20. J Proteomics. 2022. PMID: 35065287 Review.
-
Metabolic regulation of gene expression through histone acylations.Nat Rev Mol Cell Biol. 2017 Feb;18(2):90-101. doi: 10.1038/nrm.2016.140. Epub 2016 Dec 7. Nat Rev Mol Cell Biol. 2017. PMID: 27924077 Free PMC article. Review.
Cited by
-
Epigenetics Identifier screens reveal regulators of chromatin acylation and limited specificity of acylation antibodies.Sci Rep. 2021 Jun 17;11(1):12795. doi: 10.1038/s41598-021-91359-0. Sci Rep. 2021. PMID: 34140538 Free PMC article.
-
Histone Acetylation Dynamics Integrates Metabolic Activity to Regulate Plant Response to Stress.Front Plant Sci. 2019 Oct 4;10:1236. doi: 10.3389/fpls.2019.01236. eCollection 2019. Front Plant Sci. 2019. PMID: 31636650 Free PMC article. Review.
-
iKcr_CNN: A novel computational tool for imbalance classification of human nonhistone crotonylation sites based on convolutional neural networks with focal loss.Comput Struct Biotechnol J. 2022 Jun 16;20:3268-3279. doi: 10.1016/j.csbj.2022.06.032. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35832615 Free PMC article.
-
Inhibition of ACSS2-mediated histone crotonylation alleviates kidney fibrosis via IL-1β-dependent macrophage activation and tubular cell senescence.Nat Commun. 2024 Apr 13;15(1):3200. doi: 10.1038/s41467-024-47315-3. Nat Commun. 2024. PMID: 38615014 Free PMC article.
-
An Epigenetic Alphabet of Crop Adaptation to Climate Change.Front Genet. 2022 Feb 16;13:818727. doi: 10.3389/fgene.2022.818727. eCollection 2022. Front Genet. 2022. PMID: 35251130 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

