Post-translational Modification-Based Regulation of HIV Replication
- PMID: 30254620
- PMCID: PMC6141784
- DOI: 10.3389/fmicb.2018.02131
Post-translational Modification-Based Regulation of HIV Replication
Abstract
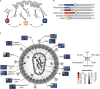
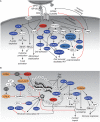
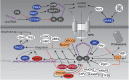
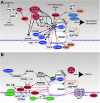
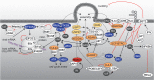
Human immunodeficiency virus (HIV) relies heavily on the host cellular machinery for production of viral progeny. To exploit cellular proteins for replication and to overcome host factors with antiviral activity, HIV has evolved a set of regulatory and accessory proteins to shape an optimized environment for its replication and to facilitate evasion from the immune system. Several cellular pathways are hijacked by the virus to modulate critical steps during the viral life cycle. Thereby, post-translational modifications (PTMs) of viral and cellular proteins gain increasingly attention as modifying enzymes regulate virtually every step of the viral replication cycle. This review summarizes the current knowledge of HIV-host interactions influenced by PTMs with a special focus on acetylation, ubiquitination, and phosphorylation of proteins linked to cellular signaling and viral replication. Insights into these interactions are surmised to aid development of new intervention strategies.
Keywords: HIV; HIV life cycle; PTM; post-translational modification; viral replication.
Figures
Similar articles
-
Role of Host-Mediated Post-Translational Modifications (PTMs) in RNA Virus Pathogenesis.Int J Mol Sci. 2020 Dec 30;22(1):323. doi: 10.3390/ijms22010323. Int J Mol Sci. 2020. PMID: 33396899 Free PMC article. Review.
-
Role of Post-translational Modifications in Influenza A Virus Life Cycle and Host Innate Immune Response.Front Microbiol. 2020 Sep 4;11:517461. doi: 10.3389/fmicb.2020.517461. eCollection 2020. Front Microbiol. 2020. PMID: 33013775 Free PMC article. Review.
-
Post-translational modifications inducing proteasomal degradation to counter HIV-1 infection.Virus Res. 2020 Nov;289:198142. doi: 10.1016/j.virusres.2020.198142. Epub 2020 Aug 31. Virus Res. 2020. PMID: 32882242 Review.
-
Lysine-specific post-translational modifications of proteins in the life cycle of viruses.Cell Cycle. 2019 Sep;18(17):1995-2005. doi: 10.1080/15384101.2019.1639305. Epub 2019 Jul 10. Cell Cycle. 2019. PMID: 31291816 Free PMC article. Review.
-
Dual role of host cell factors in HIV-1 replication: restriction and enhancement of the viral cycle.AIDS Rev. 2010 Apr-Jun;12(2):103-12. AIDS Rev. 2010. PMID: 20571604 Review.
Cited by
-
Post-Translational Modifications of Retroviral HIV-1 Gag Precursors: An Overview of Their Biological Role.Int J Mol Sci. 2021 Mar 11;22(6):2871. doi: 10.3390/ijms22062871. Int J Mol Sci. 2021. PMID: 33799890 Free PMC article. Review.
-
Mono-ADP-ribosylation by PARP10 inhibits Chikungunya virus nsP2 proteolytic activity and viral replication.Cell Mol Life Sci. 2023 Feb 25;80(3):72. doi: 10.1007/s00018-023-04717-8. Cell Mol Life Sci. 2023. PMID: 36840772 Free PMC article.
-
FusionAI: Predicting fusion breakpoint from DNA sequence with deep learning.iScience. 2021 Sep 25;24(10):103164. doi: 10.1016/j.isci.2021.103164. eCollection 2021 Oct 22. iScience. 2021. PMID: 34646994 Free PMC article.
-
Epigenetic Compound Screening Uncovers Small Molecules for Reactivation of Latent HIV-1.Antimicrob Agents Chemother. 2020 Dec 16;65(1):e01815-20. doi: 10.1128/AAC.01815-20. Print 2020 Dec 16. Antimicrob Agents Chemother. 2020. PMID: 33139279 Free PMC article.
-
In silico prediction of HIV-1-host molecular interactions and their directionality.PLoS Comput Biol. 2022 Feb 8;18(2):e1009720. doi: 10.1371/journal.pcbi.1009720. eCollection 2022 Feb. PLoS Comput Biol. 2022. PMID: 35134057 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

