Combination therapy with anti-HIV-1 antibodies maintains viral suppression
- PMID: 30258136
- PMCID: PMC6166473
- DOI: 10.1038/s41586-018-0531-2
Combination therapy with anti-HIV-1 antibodies maintains viral suppression
Abstract
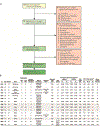
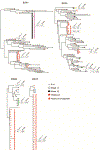
Individuals infected with HIV-1 require lifelong antiretroviral therapy, because interruption of treatment leads to rapid rebound viraemia. Here we report on a phase 1b clinical trial in which a combination of 3BNC117 and 10-1074, two potent monoclonal anti-HIV-1 broadly neutralizing antibodies that _target independent sites on the HIV-1 envelope spike, was administered during analytical treatment interruption. Participants received three infusions of 30 mg kg-1 of each antibody at 0, 3 and 6 weeks. Infusions of the two antibodies were generally well-tolerated. The nine enrolled individuals with antibody-sensitive latent viral reservoirs maintained suppression for between 15 and more than 30 weeks (median of 21 weeks), and none developed viruses that were resistant to both antibodies. We conclude that the combination of the anti-HIV-1 monoclonal antibodies 3BNC117 and 10-1074 can maintain long-term suppression in the absence of antiretroviral therapy in individuals with antibody-sensitive viral reservoirs.
Figures

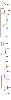
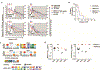



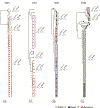




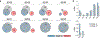
Comment in
-
Antibodies pose a double threat to HIV.Nature. 2018 Sep;561(7724):468-470. doi: 10.1038/d41586-018-06773-8. Nature. 2018. PMID: 30258147 No abstract available.
-
Long-lasting HIV suppression by combined immunotherapy.Lancet HIV. 2018 Dec;5(12):e680. doi: 10.1016/S2352-3018(18)30294-7. Lancet HIV. 2018. PMID: 30527324 No abstract available.
Similar articles
-
HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption.Nature. 2016 Jul 28;535(7613):556-60. doi: 10.1038/nature18929. Epub 2016 Jun 22. Nature. 2016. PMID: 27338952 Free PMC article. Clinical Trial.
-
Combination anti-HIV antibodies provide sustained virological suppression.Nature. 2022 Jun;606(7913):375-381. doi: 10.1038/s41586-022-04797-9. Epub 2022 Jun 1. Nature. 2022. PMID: 35650437 Free PMC article. Clinical Trial.
-
Viraemia suppressed in HIV-1-infected humans by broadly neutralizing antibody 3BNC117.Nature. 2015 Jun 25;522(7557):487-91. doi: 10.1038/nature14411. Epub 2015 Apr 8. Nature. 2015. PMID: 25855300 Free PMC article. Clinical Trial.
-
Broadly neutralizing anti-HIV-1 monoclonal antibodies in the clinic.Nat Med. 2019 Apr;25(4):547-553. doi: 10.1038/s41591-019-0412-8. Epub 2019 Apr 1. Nat Med. 2019. PMID: 30936546 Free PMC article. Review.
-
Antiviral Therapy by HIV-1 Broadly Neutralizing and Inhibitory Antibodies.Int J Mol Sci. 2016 Nov 18;17(11):1901. doi: 10.3390/ijms17111901. Int J Mol Sci. 2016. PMID: 27869733 Free PMC article. Review.
Cited by
-
Epitope-_targeting platform for broadly protective influenza vaccines.PLoS One. 2021 May 27;16(5):e0252170. doi: 10.1371/journal.pone.0252170. eCollection 2021. PLoS One. 2021. PMID: 34043704 Free PMC article.
-
Long-acting approaches for delivery of antiretroviral drugs for prevention and treatment of HIV: a review of recent research.Expert Opin Drug Deliv. 2020 Sep;17(9):1227-1238. doi: 10.1080/17425247.2020.1783233. Epub 2020 Jul 6. Expert Opin Drug Deliv. 2020. PMID: 32552187 Free PMC article. Review.
-
Influenza virus glycoprotein-reactive human monoclonal antibodies.Microbes Infect. 2020 Jul-Aug;22(6-7):263-271. doi: 10.1016/j.micinf.2020.06.003. Epub 2020 Jun 19. Microbes Infect. 2020. PMID: 32569735 Free PMC article. Review.
-
Monoclonal Antibodies as Long-Acting Products: What Are We Learning From Human Immunodeficiency Virus (HIV) and Coronavirus Disease 2019 (COVID-19)?Clin Infect Dis. 2022 Nov 21;75(Suppl 4):S530-S540. doi: 10.1093/cid/ciac751. Clin Infect Dis. 2022. PMID: 36410387 Free PMC article.
-
Initiation of Antiretroviral Therapy during Primary HIV Infection: Effects on the Latent HIV Reservoir, Including on Analytic Treatment Interruptions.AIDS Rev. 2020 Oct 26;23(1):28-39. doi: 10.24875/AIDSRev.20000001. AIDS Rev. 2020. PMID: 33105471 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

