Comparative genomic analysis of embryonic, lineage-converted and stem cell-derived motor neurons
- PMID: 30337375
- PMCID: PMC6262794
- DOI: 10.1242/dev.168617
Comparative genomic analysis of embryonic, lineage-converted and stem cell-derived motor neurons
Abstract
Advances in stem cell science allow the production of different cell types in vitro either through the recapitulation of developmental processes, often termed 'directed differentiation', or the forced expression of lineage-specific transcription factors. Although cells produced by both approaches are increasingly used in translational applications, their quantitative similarity to their primary counterparts remains largely unresolved. To investigate the similarity between in vitro-derived and primary cell types, we harvested and purified mouse spinal motor neurons and compared them with motor neurons produced by transcription factor-mediated lineage conversion of fibroblasts or directed differentiation of pluripotent stem cells. To enable unbiased analysis of these motor neuron types and their cells of origin, we then subjected them to whole transcriptome and DNA methylome analysis by RNA sequencing (RNA-seq) and reduced representation bisulfite sequencing (RRBS). Despite major differences in methodology, lineage conversion and directed differentiation both produce cells that closely approximate the primary motor neuron state. However, we identify differences in Fas signaling, the Hox code and synaptic gene expression between lineage-converted and directed differentiation motor neurons that affect their utility in translational studies.
Keywords: Directed differentiation; Embryonic stem cells; Lineage conversion; Motor neuron; Reprogramming; iPS cell.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsJ.K.I. is a co-founder of AcuraStem. K.E. is a co-founder of Q-State Biosciences and QurAlis.
Figures
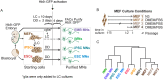


Similar articles
-
How to make spinal motor neurons.Development. 2014 Feb;141(3):491-501. doi: 10.1242/dev.097410. Development. 2014. PMID: 24449832 Review.
-
Dlk1-Dio3 locus-derived lncRNAs perpetuate postmitotic motor neuron cell fate and subtype identity.Elife. 2018 Oct 12;7:e38080. doi: 10.7554/eLife.38080. Elife. 2018. PMID: 30311912 Free PMC article.
-
Directed differentiation of human-induced pluripotent stem cells generates active motor neurons.Stem Cells. 2009 Apr;27(4):806-11. doi: 10.1002/stem.31. Stem Cells. 2009. PMID: 19350680 Free PMC article.
-
Motor neuron differentiation from pluripotent stem cells and other intermediate proliferative precursors that can be discriminated by lineage specific reporters.Stem Cell Rev Rep. 2015 Feb;11(1):194-204. doi: 10.1007/s12015-014-9541-0. Stem Cell Rev Rep. 2015. PMID: 25091426
-
Lineage choice and differentiation in mouse embryos and embryonic stem cells.Dev Biol. 2003 Dec 1;264(1):1-14. doi: 10.1016/s0012-1606(03)00390-7. Dev Biol. 2003. PMID: 14623228 Review.
Cited by
-
Proliferation history and transcription factor levels drive direct conversion.bioRxiv [Preprint]. 2023 Nov 27:2023.11.26.568736. doi: 10.1101/2023.11.26.568736. bioRxiv. 2023. PMID: 38077004 Free PMC article. Preprint.
-
A critical look: Challenges in differentiating human pluripotent stem cells into desired cell types and organoids.Wiley Interdiscip Rev Dev Biol. 2020 May;9(3):e368. doi: 10.1002/wdev.368. Epub 2019 Nov 19. Wiley Interdiscip Rev Dev Biol. 2020. PMID: 31746148 Free PMC article. Review.
-
Neuromuscular disease modeling on a chip.Dis Model Mech. 2020 Jul 7;13(7):dmm044867. doi: 10.1242/dmm.044867. Dis Model Mech. 2020. PMID: 32817118 Free PMC article. Review.
-
Canalizing cell fate by transcriptional repression.Mol Syst Biol. 2024 Mar;20(3):144-161. doi: 10.1038/s44320-024-00014-z. Epub 2024 Feb 1. Mol Syst Biol. 2024. PMID: 38302581 Free PMC article. Review.
-
Evolving principles underlying neural lineage conversion and their relevance for biomedical translation.F1000Res. 2019 Aug 30;8:F1000 Faculty Rev-1548. doi: 10.12688/f1000research.18926.1. eCollection 2019. F1000Res. 2019. PMID: 31559012 Free PMC article. Review.
References
-
- Abernathy D. G., Kim W. K., McCoy M. J., Lake A. M., Ouwenga R., Lee S. W., Xing X., Li D., Lee H. J., Heuckeroth R. O. et al. (2017). MicroRNAs induce a permissive chromatin environment that enables neuronal subtype-specific reprogramming of adult human fibroblasts. Cell Stem Cell 21, 332-348.e339. 10.1016/j.stem.2017.08.002 - DOI - PMC - PubMed
-
- Bindea G., Mlecnik B., Hackl H., Charoentong P., Tosolini M., Kirilovsky A., Fridman W.-H., Pagès F., Trajanoski Z. and Galon J. (2009). ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091-1093. 10.1093/bioinformatics/btp101 - DOI - PMC - PubMed
-
- Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z. D., Ziller M., Croft G. F., Amoroso M. W., Oakley D. H. et al. (2011). Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144, 439-452. 10.1016/j.cell.2010.12.032 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

